Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 116295 |
| Name | oriT_pAt2 |
| Organism | Agrobacterium tumefaciens strain Gle002 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP048568 (69001..69040 [+], 40 nt) |
| oriT length | 40 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 40 nt
>oriT_pAt2
TCCAAGGGCGCAATTATACGTCGCTGACACGACGCCTTGC
TCCAAGGGCGCAATTATACGTCGCTGACACGACGCCTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file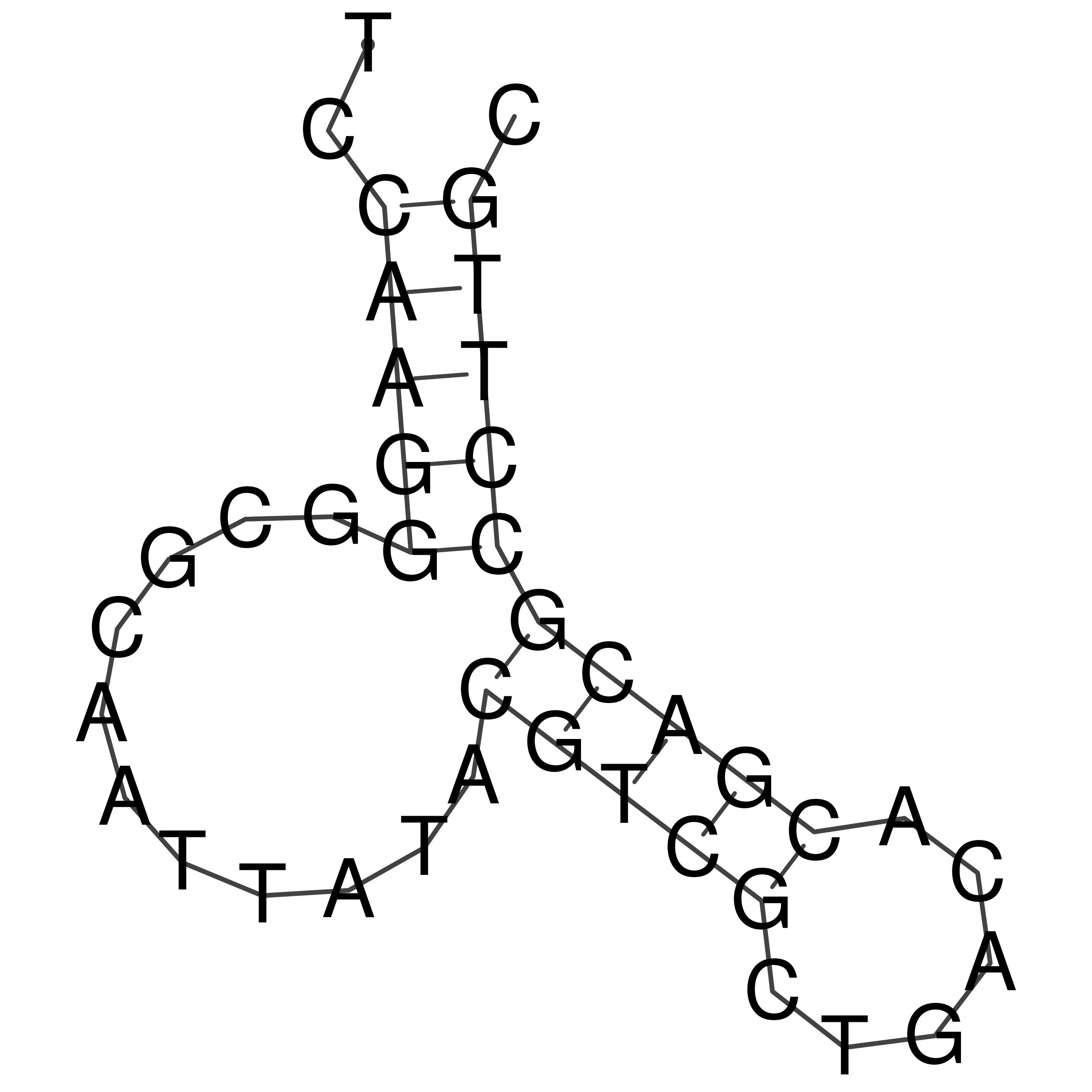
Relaxase
| ID | 10419 | GenBank | WP_174113225 |
| Name | traA_FY133_RS27100_pAt2 |
UniProt ID | _ |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 123770.52 Da Isoelectric Point: 9.9361
>WP_174113225.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Agrobacterium]
MAIAHFSASIVSRGGGRSVVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFVLPIDAPKWARAMIADR
SVSGAVEAFWNKVEDFEKRSDAQLARDLTIALPLELTPEQNIALVRDFVEKHIHAKGMVADWVYHNNPGN
PHIHLMTTLRPLTEDGFGAKKVAVTAKDGQPVRTKSGKILYELWAGSTDDFNVLRDGWFERLNHHLALGG
IDLRIDGRSYEKQGIDLEPAIHLGVGAKAIERKAQEQGFRPELERLELNEERRGENARRILRRPEIVLDL
IMREKSVFDERDVAKVLHRYIDDPAVFQQLMLRIILNPEVLRLQRDTIDFSTGEKVPARYSTRAMIRLEA
TMVRQATWLSGKDTHAVSAAVLDATLRRHERLSEEQKTAIERIAGPARIAAVVGRAGAGKTTMMKAVREA
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWNRGRDVLDSKTIFVMDEAGMVASKQMAGFVD
TVVRAGAKIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALTAYD
ANVRITGTRLKAEAVERLIGDWNHDYDHTKTTLILAHLRRDVRMLNQMAREKLVERGIVGEGHVFKTADG
IRQFDAGDQIVFLKNEGSLGVKNGMIAHIIEAAPNRIVAVIGEGDQRRRVIVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGRRSFAFNGGLSKVLSRKNAKETTLDYERGSLY
REALRYAENRGLNIVQVARTLLRDQLDWTLRQKTRLDELGQRLAVFAERLGLHHPPKTQTMKEAAPMVAG
IKTFPGSVADTVGDRLGTDPALKRQWEEVSTRFAYVFADPETAFRAMNFDAVLADKDSALQVLQKLEAEP
ASIGPLKGKTGILASKTDREDRRIAELNVPALKRDLEQYLRMRENAAQRLQTDEQALRQRVSIDIPALSP
AARVVLERVRDAIDRNDLPAALGYVLSNRETKLEIDGFSKAVTERFGERTLLTNAARQPSGPLFDKLSNG
MRPAQKEQLKEAWPVMRTAQQIAAHERTTQTLKQAEELRLTQRQSPVMKQ
MAIAHFSASIVSRGGGRSVVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFVLPIDAPKWARAMIADR
SVSGAVEAFWNKVEDFEKRSDAQLARDLTIALPLELTPEQNIALVRDFVEKHIHAKGMVADWVYHNNPGN
PHIHLMTTLRPLTEDGFGAKKVAVTAKDGQPVRTKSGKILYELWAGSTDDFNVLRDGWFERLNHHLALGG
IDLRIDGRSYEKQGIDLEPAIHLGVGAKAIERKAQEQGFRPELERLELNEERRGENARRILRRPEIVLDL
IMREKSVFDERDVAKVLHRYIDDPAVFQQLMLRIILNPEVLRLQRDTIDFSTGEKVPARYSTRAMIRLEA
TMVRQATWLSGKDTHAVSAAVLDATLRRHERLSEEQKTAIERIAGPARIAAVVGRAGAGKTTMMKAVREA
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWNRGRDVLDSKTIFVMDEAGMVASKQMAGFVD
TVVRAGAKIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALTAYD
ANVRITGTRLKAEAVERLIGDWNHDYDHTKTTLILAHLRRDVRMLNQMAREKLVERGIVGEGHVFKTADG
IRQFDAGDQIVFLKNEGSLGVKNGMIAHIIEAAPNRIVAVIGEGDQRRRVIVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGRRSFAFNGGLSKVLSRKNAKETTLDYERGSLY
REALRYAENRGLNIVQVARTLLRDQLDWTLRQKTRLDELGQRLAVFAERLGLHHPPKTQTMKEAAPMVAG
IKTFPGSVADTVGDRLGTDPALKRQWEEVSTRFAYVFADPETAFRAMNFDAVLADKDSALQVLQKLEAEP
ASIGPLKGKTGILASKTDREDRRIAELNVPALKRDLEQYLRMRENAAQRLQTDEQALRQRVSIDIPALSP
AARVVLERVRDAIDRNDLPAALGYVLSNRETKLEIDGFSKAVTERFGERTLLTNAARQPSGPLFDKLSNG
MRPAQKEQLKEAWPVMRTAQQIAAHERTTQTLKQAEELRLTQRQSPVMKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 12164 | GenBank | WP_153517022 |
| Name | traG_FY133_RS27085_pAt2 |
UniProt ID | _ |
| Length | 641 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 641 a.a. Molecular weight: 69244.33 Da Isoelectric Point: 9.4596
>WP_153517022.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Agrobacterium]
MTVNRLLLLILPAIIMLAAMFATSGTEQRLAAFGTSPQAKLMLGRAGLALPYIAAVAIGLIGLFAANGSA
NIKAAGLSVLAGNGVVITIATLREVIRLNSITSRVPAEQSVLAYADPVTMIGVGIAFISGMFALRVAIKG
NAAFATTAPKRIGGKRAVHGEADWMKIQEAAKLFPEPGGIVIGERYRVDRDSVATMPFRADDRQSWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSSEVAPMVYEHRRQAGRKVIVLD
PTAGGVGFNALDWIGRHGSTKEEDIVAVATWIMTDNPRTASARDDFFRASAMQLLTALIADVCLSGHTEA
EDQTLRRVRANLSEPEPKLRARLTKIYEGSESDFVKENVSVFVNMTPETFSGVYANAVKETHWLSYPNYA
GLVSGDSFSTDDLADGGTDIFIALDLKVLEAHPGLARVVIGSLLNAIYNRNGDVKGRTLFLLDEVARLGY
LRILETARDAGRKYGTTLTMIFQSIGQMREAYGGRDATSKWFESASWISFAAINDPDTADYISKRCGDTT
VEVDQTNRSSGMKGSSRSRSRQLSRRPLILPHEVLRMRADEQIVFTSGNPPLRCGRAIWFRRDDMKACVG
TNRFHREEKAP
MTVNRLLLLILPAIIMLAAMFATSGTEQRLAAFGTSPQAKLMLGRAGLALPYIAAVAIGLIGLFAANGSA
NIKAAGLSVLAGNGVVITIATLREVIRLNSITSRVPAEQSVLAYADPVTMIGVGIAFISGMFALRVAIKG
NAAFATTAPKRIGGKRAVHGEADWMKIQEAAKLFPEPGGIVIGERYRVDRDSVATMPFRADDRQSWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSSEVAPMVYEHRRQAGRKVIVLD
PTAGGVGFNALDWIGRHGSTKEEDIVAVATWIMTDNPRTASARDDFFRASAMQLLTALIADVCLSGHTEA
EDQTLRRVRANLSEPEPKLRARLTKIYEGSESDFVKENVSVFVNMTPETFSGVYANAVKETHWLSYPNYA
GLVSGDSFSTDDLADGGTDIFIALDLKVLEAHPGLARVVIGSLLNAIYNRNGDVKGRTLFLLDEVARLGY
LRILETARDAGRKYGTTLTMIFQSIGQMREAYGGRDATSKWFESASWISFAAINDPDTADYISKRCGDTT
VEVDQTNRSSGMKGSSRSRSRQLSRRPLILPHEVLRMRADEQIVFTSGNPPLRCGRAIWFRRDDMKACVG
TNRFHREEKAP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 12165 | GenBank | WP_080868325 |
| Name | t4cp2_FY133_RS27435_pAt2 |
UniProt ID | _ |
| Length | 822 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 822 a.a. Molecular weight: 92115.82 Da Isoelectric Point: 5.6807
>WP_080868325.1 MULTISPECIES: conjugal transfer protein TrbE [Rhizobium/Agrobacterium group]
MVALKLFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDSERNEVSRQINMIMSRLG
SGWMVQVEALRVPTIDYPVGDTCHFPDPVTRAIDTERRSHFERESGHFESRHALILTWRPPEPRRSGLAR
YVYSDTASRSATYADTALDSFRTSIREFEQYLANVVSIRRMVTRETEERGGFRVARYDELFQFIRFCITG
ENHPVRLPDIPMYLDWLVTAELQHGLTPTVENRFLGVVAIDGLAAESWPGILNSLDLMPLTYRWSSRFVF
LDEQEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMMMVAETEDAIAEASSQLVAYGYYTPVILL
FDEQQERLQEKCEAIRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLVNTRNLADLIPLNSVWS
GSPVAPCPFYPPGSPPLMQVASGSTPFRLNLHVDDVGHTLVFGPTGSGKSTLLSLIAAQFRRYADAQIFA
FDKGRSMLPLTRAIGGDHYEIGGDTAEGGEGASPQLAFCPLAELASDSDRAWAAEWIETLVALQGVTVTP
DYRNAISRQLGLMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEQDGLALGAFQCFEVEEL
MNMGERNLVPVLLYMFRRVEKRLTGAPSLIILDEAWLMLGHPTFRDKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATALPKREYYVASPEGRRLFDM
SLGPVALSFVGASGKEDLKRIRALHFQHGADWPKHWLQQRGIAHAETLFQAA
MVALKLFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDSERNEVSRQINMIMSRLG
SGWMVQVEALRVPTIDYPVGDTCHFPDPVTRAIDTERRSHFERESGHFESRHALILTWRPPEPRRSGLAR
YVYSDTASRSATYADTALDSFRTSIREFEQYLANVVSIRRMVTRETEERGGFRVARYDELFQFIRFCITG
ENHPVRLPDIPMYLDWLVTAELQHGLTPTVENRFLGVVAIDGLAAESWPGILNSLDLMPLTYRWSSRFVF
LDEQEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMMMVAETEDAIAEASSQLVAYGYYTPVILL
FDEQQERLQEKCEAIRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLVNTRNLADLIPLNSVWS
GSPVAPCPFYPPGSPPLMQVASGSTPFRLNLHVDDVGHTLVFGPTGSGKSTLLSLIAAQFRRYADAQIFA
FDKGRSMLPLTRAIGGDHYEIGGDTAEGGEGASPQLAFCPLAELASDSDRAWAAEWIETLVALQGVTVTP
DYRNAISRQLGLMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEQDGLALGAFQCFEVEEL
MNMGERNLVPVLLYMFRRVEKRLTGAPSLIILDEAWLMLGHPTFRDKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATALPKREYYVASPEGRRLFDM
SLGPVALSFVGASGKEDLKRIRALHFQHGADWPKHWLQQRGIAHAETLFQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 137949..147619
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| FY133_RS27375 (FY133_27150) | 133469..134194 | - | 726 | WP_080868320 | nopaline ABC transporter permease NocM | - |
| FY133_RS27380 (FY133_27155) | 134196..134906 | - | 711 | WP_080868351 | nopaline ABC transporter permease NocQ | - |
| FY133_RS27385 (FY133_27160) | 134979..135830 | - | 852 | WP_003523898 | nopaline ABC transporter substrate-binding protein NocT | - |
| FY133_RS27390 (FY133_27165) | 135895..136668 | - | 774 | WP_080868321 | nopaline ABC transporter ATP-binding protein NocP | - |
| FY133_RS27395 (FY133_27170) | 136918..137820 | + | 903 | WP_045023359 | LysR family transcriptional regulator | - |
| FY133_RS27400 (FY133_27175) | 137949..139256 | - | 1308 | WP_080868322 | IncP-type conjugal transfer protein TrbI | virB10 |
| FY133_RS27405 (FY133_27180) | 139271..139747 | - | 477 | WP_045025162 | conjugal transfer protein TrbH | - |
| FY133_RS27410 (FY133_27185) | 139747..140601 | - | 855 | WP_070167554 | P-type conjugative transfer protein TrbG | virB9 |
| FY133_RS27415 (FY133_27190) | 140619..141281 | - | 663 | WP_080868323 | conjugal transfer protein TrbF | virB8 |
| FY133_RS27420 (FY133_27195) | 141288..142490 | - | 1203 | WP_080868324 | P-type conjugative transfer protein TrbL | virB6 |
| FY133_RS27425 (FY133_27200) | 142484..142702 | - | 219 | WP_070167519 | entry exclusion protein TrbK | - |
| FY133_RS27430 (FY133_27205) | 142699..143508 | - | 810 | WP_070167518 | P-type conjugative transfer protein TrbJ | virB5 |
| FY133_RS27435 (FY133_27210) | 143480..145948 | - | 2469 | WP_080868325 | conjugal transfer protein TrbE | virb4 |
| FY133_RS27440 (FY133_27215) | 145959..146258 | - | 300 | WP_080868326 | conjugal transfer protein TrbD | virB3 |
| FY133_RS27445 (FY133_27220) | 146251..146658 | - | 408 | WP_080868327 | conjugal transfer pilin TrbC | virB2 |
| FY133_RS27450 (FY133_27225) | 146648..147619 | - | 972 | WP_080868328 | P-type conjugative transfer ATPase TrbB | virB11 |
| FY133_RS27455 (FY133_27230) | 147616..148251 | - | 636 | WP_041723972 | acyl-homoserine-lactone synthase | - |
Host bacterium
| ID | 16728 | GenBank | NZ_CP048568 |
| Plasmid name | pAt2 | Incompatibility group | - |
| Plasmid size | 148616 bp | Coordinate of oriT [Strand] | 69001..69040 [+] |
| Host baterium | Agrobacterium tumefaciens strain Gle002 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |