Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 116265 |
| Name | oriT_ES-1390|unnamed1 |
| Organism | Enterococcus faecalis strain ES-1390 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP136354 (38610..38659 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_ES-1390|unnamed1
AATATCGCAACATGCTAGCATGTTGCTCTGCTCGCAAAAAGAAAGCCTAC
AATATCGCAACATGCTAGCATGTTGCTCTGCTCGCAAAAAGAAAGCCTAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file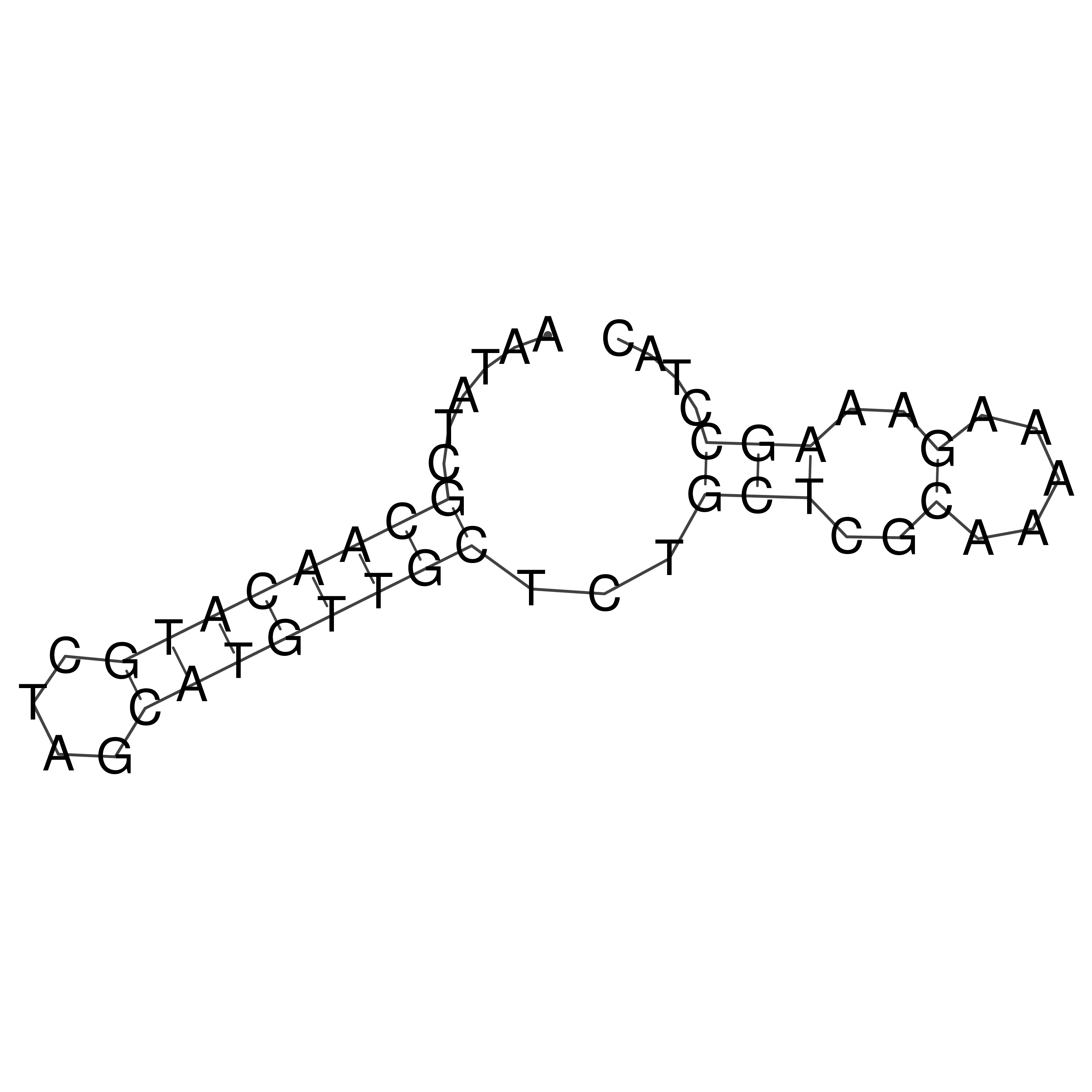
Relaxase
| ID | 10402 | GenBank | WP_002367932 |
| Name | Relaxase_RX141_RS14005_ES-1390|unnamed1 |
UniProt ID | R3K157 |
| Length | 561 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 561 a.a. Molecular weight: 65400.15 Da Isoelectric Point: 6.0174
>WP_002367932.1 MULTISPECIES: relaxase/mobilization nuclease domain-containing protein [Enterococcus]
MVYTKHFVIHTFDKLNNACSYIENAEKTEVTNDNPSEHLEHLFQYIVNDDKTYMKKLVSGHGIVDPTNPY
EEFKLTKLQAAIQRKIGYTFDPKSERLLPPTLTELEKGNAVLAHHLIQSFSPEDDLTPEKIHEIGYNTVM
ELTGGEYEFVIATHVDKEHLHNHIIFSSTNLKTGKAFRWQKGTKKVFEQISDKIAAKEGAKIIEKSPKNS
HKKYTMWETESLYKEKIKSRLDFLLEQSSSINDFLEKAEALNLSVDFSKKWTTYRLLDEPQIKNTRSRSL
SKSDPTRYNYEKIVERLKENKSVLTLKEVVEKYVEKSEKSKNEFDYQLTIDEWQISHKTTRGYYVNVDFG
FGERGKVFIGAYKVDPLENGQYNIYVKRKDFFYLMNEKNADRNRYMTGETLIKQLRLYNGQTPLKKEPVM
RTIDELVNAINFLAANEIEDTRQLELLEEKLEAAFLEAEETLETLDEKMLELHQLSNLLLENELNGELEL
IQQKLKTLLPDATLAEFSYEDVRGEIEAIKTSQSLLENKLERTRNEINQLHEIQAVQKKETANQNNVKPK
L
MVYTKHFVIHTFDKLNNACSYIENAEKTEVTNDNPSEHLEHLFQYIVNDDKTYMKKLVSGHGIVDPTNPY
EEFKLTKLQAAIQRKIGYTFDPKSERLLPPTLTELEKGNAVLAHHLIQSFSPEDDLTPEKIHEIGYNTVM
ELTGGEYEFVIATHVDKEHLHNHIIFSSTNLKTGKAFRWQKGTKKVFEQISDKIAAKEGAKIIEKSPKNS
HKKYTMWETESLYKEKIKSRLDFLLEQSSSINDFLEKAEALNLSVDFSKKWTTYRLLDEPQIKNTRSRSL
SKSDPTRYNYEKIVERLKENKSVLTLKEVVEKYVEKSEKSKNEFDYQLTIDEWQISHKTTRGYYVNVDFG
FGERGKVFIGAYKVDPLENGQYNIYVKRKDFFYLMNEKNADRNRYMTGETLIKQLRLYNGQTPLKKEPVM
RTIDELVNAINFLAANEIEDTRQLELLEEKLEAAFLEAEETLETLDEKMLELHQLSNLLLENELNGELEL
IQQKLKTLLPDATLAEFSYEDVRGEIEAIKTSQSLLENKLERTRNEINQLHEIQAVQKKETANQNNVKPK
L
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | R3K157 |
Auxiliary protein
| ID | 6114 | GenBank | WP_002362408 |
| Name | WP_002362408_ES-1390|unnamed1 |
UniProt ID | A0A1J6XY92 |
| Length | 118 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 118 a.a. Molecular weight: 13979.04 Da Isoelectric Point: 9.6545
>WP_002362408.1 MULTISPECIES: plasmid mobilization relaxosome protein MobC [Bacillota]
MENKQKLKRPIQRIVRLSEEENNLIKRKIEESFFPNFQNFALHLLIQGEIRHVDYSELNRLTTEIHKIGI
NINQMARLANQFHEISSEDIKDLTDKVQSLNALVQSELNKLIKRKEQS
MENKQKLKRPIQRIVRLSEEENNLIKRKIEESFFPNFQNFALHLLIQGEIRHVDYSELNRLTTEIHKIGI
NINQMARLANQFHEISSEDIKDLTDKVQSLNALVQSELNKLIKRKEQS
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A1J6XY92 |
T4CP
| ID | 12142 | GenBank | WP_316498578 |
| Name | t4cp2_RX141_RS14025_ES-1390|unnamed1 |
UniProt ID | _ |
| Length | 609 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 609 a.a. Molecular weight: 70072.04 Da Isoelectric Point: 6.0617
>WP_316498578.1 VirD4-like conjugal transfer protein, CD1115 family [Enterococcus faecalis]
MQTYKKELKPYLYLGVFLFVIGFGIGNFFWLLPGTDYFSKTNYLFTKDILTYIEQTFYRFFITPSLLGFS
VGVLGFLLGLLMYVRDNDRGIYRHGEEYGSARFATPAEMKKYEDPIPENNIIVSKHVQISLFNKRLPIKL
QKNKNIAILGDSGAAKTLAFIKTNLMQRHASFITTDPDGGILPEIGLLLKKGKYKIKVLDLNTLSNSDTF
NVFEYMHSELDVDRVLEAVTEATKEVEKTTGDDFWIKAEGLLVRSLIAYLWFDGRRNDYTPHLGMIADML
RHLKRKDKNVPSPAEEWFEELNEAIPDNYAYRQWTLFNNLYESETRASVLGIAAARYSVFDHEEVVNMIR
TDTMDIDSWNEEKTAVFIAIPETNTSFNFIAALMFATVGEVLRNKSDQVRLGKRVLEEGKELLHVRFLID
EFANIGRIPHFEKMLTTFRKREMSFTIVLQSLNQLQTLYPKGWQNILNGCACLLYLGGDEEETTKYLSAR
AGKQTLSIRKHSMNKGNQGGGSENRDKYGRDLIDRSEVALIGGDECLVFISKEHVFKDKKFYAFQHDRAD
ELATGPTDDKWYTWRRYMTDEEAILDKISATDIIDHGEITSELKEETHV
MQTYKKELKPYLYLGVFLFVIGFGIGNFFWLLPGTDYFSKTNYLFTKDILTYIEQTFYRFFITPSLLGFS
VGVLGFLLGLLMYVRDNDRGIYRHGEEYGSARFATPAEMKKYEDPIPENNIIVSKHVQISLFNKRLPIKL
QKNKNIAILGDSGAAKTLAFIKTNLMQRHASFITTDPDGGILPEIGLLLKKGKYKIKVLDLNTLSNSDTF
NVFEYMHSELDVDRVLEAVTEATKEVEKTTGDDFWIKAEGLLVRSLIAYLWFDGRRNDYTPHLGMIADML
RHLKRKDKNVPSPAEEWFEELNEAIPDNYAYRQWTLFNNLYESETRASVLGIAAARYSVFDHEEVVNMIR
TDTMDIDSWNEEKTAVFIAIPETNTSFNFIAALMFATVGEVLRNKSDQVRLGKRVLEEGKELLHVRFLID
EFANIGRIPHFEKMLTTFRKREMSFTIVLQSLNQLQTLYPKGWQNILNGCACLLYLGGDEEETTKYLSAR
AGKQTLSIRKHSMNKGNQGGGSENRDKYGRDLIDRSEVALIGGDECLVFISKEHVFKDKKFYAFQHDRAD
ELATGPTDDKWYTWRRYMTDEEAILDKISATDIIDHGEITSELKEETHV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 41304..58590
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| RX141_RS14005 (RX141_14005) | 36501..38186 | - | 1686 | WP_002367932 | relaxase/mobilization nuclease domain-containing protein | - |
| RX141_RS14010 (RX141_14010) | 38187..38543 | - | 357 | WP_002362408 | plasmid mobilization relaxosome protein MobC | - |
| RX141_RS14015 (RX141_14015) | 38789..39061 | - | 273 | WP_002367930 | LtrD | - |
| RX141_RS14020 (RX141_14020) | 39094..41253 | - | 2160 | WP_316498577 | ArdC-like ssDNA-binding domain-containing protein | - |
| RX141_RS14025 (RX141_14025) | 41304..43133 | - | 1830 | WP_316498578 | VirD4-like conjugal transfer protein, CD1115 family | virb4 |
| RX141_RS14030 (RX141_14030) | 43133..43618 | - | 486 | WP_002386493 | PcfB family protein | gbs1365 |
| RX141_RS14035 (RX141_14035) | 43777..44385 | - | 609 | WP_316498615 | PcfA protein | - |
| RX141_RS14040 (RX141_14040) | 44369..44614 | - | 246 | WP_002365866 | hypothetical protein | - |
| RX141_RS14045 (RX141_14045) | 44601..45227 | - | 627 | WP_002386490 | hypothetical protein | prgL |
| RX141_RS14050 (RX141_14050) | 45251..47866 | - | 2616 | WP_316498581 | phage tail tip lysozyme | prgK |
| RX141_RS14055 (RX141_14055) | 47878..50268 | - | 2391 | WP_316498582 | VirB4-like conjugal transfer ATPase, CD1110 family | virb4 |
| RX141_RS14060 (RX141_14060) | 50222..50575 | - | 354 | WP_002367919 | PrgI family mobile element protein | prgIc |
| RX141_RS14065 (RX141_14065) | 50577..51377 | - | 801 | WP_002382799 | type IV secretion system protein | prgHb |
| RX141_RS14070 (RX141_14070) | 51392..51688 | - | 297 | WP_002382798 | hypothetical protein | - |
| RX141_RS14075 (RX141_14075) | 51701..51928 | - | 228 | WP_002365858 | hypothetical protein | - |
| RX141_RS14080 (RX141_14080) | 51975..52409 | - | 435 | WP_002365857 | hypothetical protein | - |
| RX141_RS14085 (RX141_14085) | 52429..53328 | - | 900 | WP_159110802 | type III secretion system protein PrgD | - |
| RX141_RS14090 (RX141_14090) | 53370..54227 | - | 858 | WP_159110801 | pheromone response LPXTG-anchored adhesin PrgC | prgC |
| RX141_RS14095 (RX141_14095) | 54255..54611 | - | 357 | WP_002367672 | pheromone response system RNA-binding regulator PrgU | - |
| RX141_RS14100 (RX141_14100) | 54673..58590 | - | 3918 | WP_316498583 | LPXTG-anchored aggregation substance PrgB/Asc10 | prgB |
| RX141_RS14105 (RX141_14105) | 58802..58990 | - | 189 | Protein_63 | hypothetical protein | - |
| RX141_RS14110 (RX141_14110) | 59263..59457 | - | 195 | WP_002415094 | hypothetical protein | - |
| RX141_RS14115 (RX141_14115) | 59536..62208 | - | 2673 | WP_316498584 | surface exclusion protein SEA1/PrgA | - |
| RX141_RS14120 (RX141_14120) | 62219..62524 | - | 306 | WP_002367666 | hypothetical protein | - |
Host bacterium
| ID | 16698 | GenBank | NZ_CP136354 |
| Plasmid name | ES-1390|unnamed1 | Incompatibility group | - |
| Plasmid size | 62970 bp | Coordinate of oriT [Strand] | 38610..38659 [-] |
| Host baterium | Enterococcus faecalis strain ES-1390 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | prgB/asc10 |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |