Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 116232 |
| Name | oriT_pSF3 |
| Organism | Enterococcus faecalis strain SF28073 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP060803 (39387..39416 [+], 30 nt) |
| oriT length | 30 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 30 nt
>oriT_pSF3
AACGTAAGTTAGTGGGTATAAGTGCGCCCT
AACGTAAGTTAGTGGGTATAAGTGCGCCCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file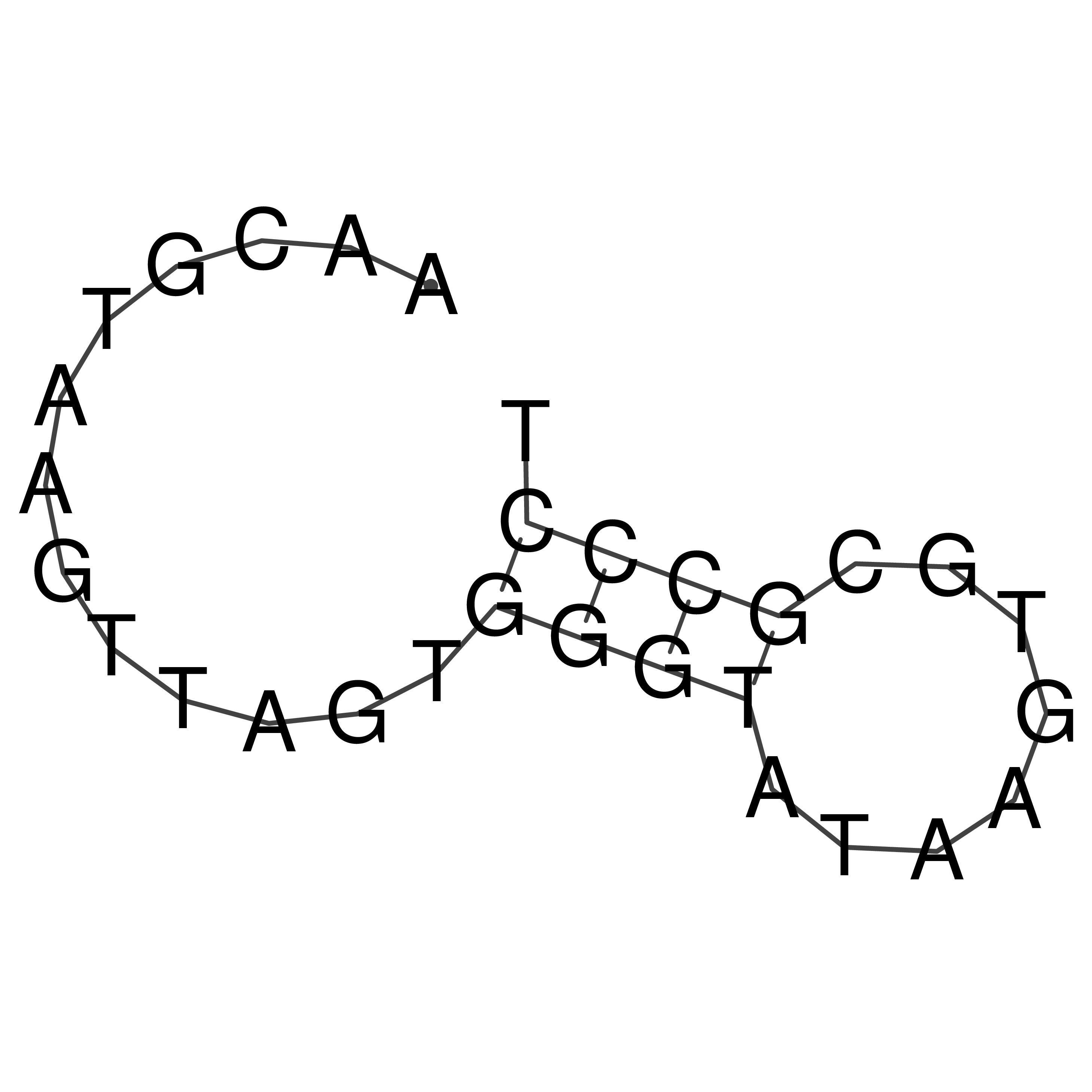
Relaxase
| ID | 10384 | GenBank | WP_000974115 |
| Name | mobQ_H9Q64_RS01025_pSF3 |
UniProt ID | A0A7H0FKC2 |
| Length | 661 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 661 a.a. Molecular weight: 77412.46 Da Isoelectric Point: 9.3565
>WP_000974115.1 MULTISPECIES: MobQ family relaxase [Bacilli]
MAIFHLSMTIAKREGGKRSLIAMASYRSGEKLYSELYEKTNLYNHRTVKPEAFILKPDYVPNEFLDRQTL
WNKMELAEKSPNAQLCREVNVALPIELNNSDQRMLIEDFVKDNFVSEGMIADVAIHRDDENNPHAHIMLT
MREVDSEGNILNKRKRIPKLDENGNQIFNEKGQRVTVSIKTNDWDRKSLVSEIRKDWADKVNQYLKDRNI
DQQITEKSHAELGKKELPTIHEGFYSKKLEDKGVISELKRKNLEIQSYNDVLSELDKLENQEKVLKQDQN
FTLKFEKTFSPLEKKELKNLSKELKIFVNDENIDKRISELKRWENSLIFNNKMEIQKQRLMLSKISSERD
MLTKANEILDKQAERFFKKSYPSLNIDKFSNHEVRAMVNETIFRKQLLNKDQLAEVIYNERVVEKEESKK
IFKEKPFQTSRYLDSKIKQVEDSITKENNPERKEILSIKKEKLIGIKQGLIEYVQSEVERKFDKNVSIDS
VIEGEMLLAKADYYKTTDFSKVEGVARFSSEEINSMLEQSKGFLTNIQTVKIPNDCQGVFFVQDSMKHID
ELSPLAKHNLKKVVNRNAYLPDSDKIELSKEIENTNKDQSQELDKDVPEKNEVTVKMFQFAKSINRLLSG
NQLQKKRNLDKLIKQTKAKENQSLQRNIPLR
MAIFHLSMTIAKREGGKRSLIAMASYRSGEKLYSELYEKTNLYNHRTVKPEAFILKPDYVPNEFLDRQTL
WNKMELAEKSPNAQLCREVNVALPIELNNSDQRMLIEDFVKDNFVSEGMIADVAIHRDDENNPHAHIMLT
MREVDSEGNILNKRKRIPKLDENGNQIFNEKGQRVTVSIKTNDWDRKSLVSEIRKDWADKVNQYLKDRNI
DQQITEKSHAELGKKELPTIHEGFYSKKLEDKGVISELKRKNLEIQSYNDVLSELDKLENQEKVLKQDQN
FTLKFEKTFSPLEKKELKNLSKELKIFVNDENIDKRISELKRWENSLIFNNKMEIQKQRLMLSKISSERD
MLTKANEILDKQAERFFKKSYPSLNIDKFSNHEVRAMVNETIFRKQLLNKDQLAEVIYNERVVEKEESKK
IFKEKPFQTSRYLDSKIKQVEDSITKENNPERKEILSIKKEKLIGIKQGLIEYVQSEVERKFDKNVSIDS
VIEGEMLLAKADYYKTTDFSKVEGVARFSSEEINSMLEQSKGFLTNIQTVKIPNDCQGVFFVQDSMKHID
ELSPLAKHNLKKVVNRNAYLPDSDKIELSKEIENTNKDQSQELDKDVPEKNEVTVKMFQFAKSINRLLSG
NQLQKKRNLDKLIKQTKAKENQSLQRNIPLR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A7H0FKC2 |
T4CP
| ID | 12122 | GenBank | WP_000096577 |
| Name | t4cp2_H9Q64_RS00980_pSF3 |
UniProt ID | _ |
| Length | 551 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 551 a.a. Molecular weight: 63042.12 Da Isoelectric Point: 4.7709
>WP_000096577.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Bacilli]
MSSLLAESDGLILGKFQSGNVVIQPEDSKISNRNIFVVGGPGSFKTQSYVLPNVVNNRSTSIVVTDPKGE
IYELTNEIKKAQGFKTVVINFKDFLLSSRYNPLLYIRKSNDTNKIANVIVSAKNDPKRKDFWFNAQLSLL
NTLIKYVYFEYEPSARTIEGILDFLEEFDPRYNEEGVSELDEQFERLPDGHEAKRSYYLGFRQAQSEARP
NIVISLLTTLQDFVDKEVSEFTSTNDFFFEELGTSKICLYVLISPLDRTWDGLVNLFFQQMFTELYFLGD
KHNAKLPVPLVMLLDEFVNLGYFPTYENFLATCRGYRISVSTILQSLPQGFELYGDKKFKAIIGNHAIKI
CLGGVEETTAEYFSRQVNDTTIKVYTGGTSESKTSAKTTNRSGSKSESYGYQKRRLITEGEVINLQQEEN
GRKSIVLIDGKPYMLRKTPQFELFGNLLKKHEISQQDYISSQTEFAKEYIEELEKQHKQRKVQLASAPIF
HEKKQEDDLNKKEINLPEQPIESEVKEEAEEKELLMSDILSSFDEGIEENTANNDNSELPF
MSSLLAESDGLILGKFQSGNVVIQPEDSKISNRNIFVVGGPGSFKTQSYVLPNVVNNRSTSIVVTDPKGE
IYELTNEIKKAQGFKTVVINFKDFLLSSRYNPLLYIRKSNDTNKIANVIVSAKNDPKRKDFWFNAQLSLL
NTLIKYVYFEYEPSARTIEGILDFLEEFDPRYNEEGVSELDEQFERLPDGHEAKRSYYLGFRQAQSEARP
NIVISLLTTLQDFVDKEVSEFTSTNDFFFEELGTSKICLYVLISPLDRTWDGLVNLFFQQMFTELYFLGD
KHNAKLPVPLVMLLDEFVNLGYFPTYENFLATCRGYRISVSTILQSLPQGFELYGDKKFKAIIGNHAIKI
CLGGVEETTAEYFSRQVNDTTIKVYTGGTSESKTSAKTTNRSGSKSESYGYQKRRLITEGEVINLQQEEN
GRKSIVLIDGKPYMLRKTPQFELFGNLLKKHEISQQDYISSQTEFAKEYIEELEKQHKQRKVQLASAPIF
HEKKQEDDLNKKEINLPEQPIESEVKEEAEEKELLMSDILSSFDEGIEENTANNDNSELPF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 24745..36921
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| H9Q64_RS00925 (H9Q64_00925) | 20231..20593 | - | 363 | WP_000108744 | toprim domain-containing protein | - |
| H9Q64_RS00930 (H9Q64_00930) | 20593..21210 | - | 618 | WP_001062588 | recombinase family protein | - |
| H9Q64_RS00935 (H9Q64_00935) | 21224..21394 | - | 171 | WP_000713595 | hypothetical protein | - |
| H9Q64_RS00940 (H9Q64_00940) | 21742..23232 | - | 1491 | WP_001025005 | primase C-terminal domain-containing protein | - |
| H9Q64_RS00945 (H9Q64_00945) | 23636..23884 | - | 249 | WP_000362415 | helix-turn-helix domain-containing protein | - |
| H9Q64_RS00950 (H9Q64_00950) | 24322..24426 | + | 105 | WP_113792438 | type I toxin-antitoxin system Fst family toxin | - |
| H9Q64_RS00955 (H9Q64_00955) | 24745..25458 | - | 714 | WP_000717317 | LPXTG cell wall anchor domain-containing protein | prgC |
| H9Q64_RS00960 (H9Q64_00960) | 25518..25886 | - | 369 | WP_000516429 | hypothetical protein | - |
| H9Q64_RS00965 (H9Q64_00965) | 25915..26883 | - | 969 | WP_000122915 | hypothetical protein | - |
| H9Q64_RS00970 (H9Q64_00970) | 26900..27832 | - | 933 | WP_000382494 | conjugal transfer protein TrbL family protein | trsL |
| H9Q64_RS00975 (H9Q64_00975) | 27834..28757 | - | 924 | WP_001013727 | hypothetical protein | traP |
| H9Q64_RS00980 (H9Q64_00980) | 28775..30430 | - | 1656 | WP_000096577 | VirD4-like conjugal transfer protein, CD1115 family | - |
| H9Q64_RS00985 (H9Q64_00985) | 30423..30854 | - | 432 | WP_001085135 | hypothetical protein | - |
| H9Q64_RS00990 (H9Q64_00990) | 30859..31410 | - | 552 | WP_000393725 | hypothetical protein | - |
| H9Q64_RS00995 (H9Q64_00995) | 31423..32532 | - | 1110 | WP_000501303 | lysozyme family protein | orf14 |
| H9Q64_RS01000 (H9Q64_01000) | 32554..33906 | - | 1353 | WP_000875579 | hypothetical protein | trsF |
| H9Q64_RS01005 (H9Q64_01005) | 33920..35881 | - | 1962 | WP_000411529 | VirB4 family type IV secretion system protein | virb4 |
| H9Q64_RS01010 (H9Q64_01010) | 35892..36521 | - | 630 | WP_000459258 | hypothetical protein | trsD |
| H9Q64_RS01015 (H9Q64_01015) | 36538..36921 | - | 384 | WP_001083514 | glycosyl transferase | trsC |
| H9Q64_RS01020 (H9Q64_01020) | 36940..37272 | - | 333 | WP_000713503 | CagC family type IV secretion system protein | - |
| H9Q64_RS01025 (H9Q64_01025) | 37296..39281 | - | 1986 | WP_000974115 | MobQ family relaxase | - |
| H9Q64_RS01030 (H9Q64_01030) | 39573..39872 | + | 300 | WP_000431202 | hypothetical protein | - |
| H9Q64_RS01035 (H9Q64_01035) | 39875..40132 | + | 258 | WP_000002668 | hypothetical protein | - |
| H9Q64_RS01040 (H9Q64_01040) | 40155..40460 | - | 306 | WP_000351790 | hypothetical protein | - |
| H9Q64_RS01045 (H9Q64_01045) | 40494..40991 | - | 498 | WP_000874092 | DnaJ domain-containing protein | - |
| H9Q64_RS01050 (H9Q64_01050) | 41030..41347 | - | 318 | WP_000415964 | hypothetical protein | - |
Host bacterium
| ID | 16665 | GenBank | NZ_CP060803 |
| Plasmid name | pSF3 | Incompatibility group | - |
| Plasmid size | 46541 bp | Coordinate of oriT [Strand] | 39387..39416 [+] |
| Host baterium | Enterococcus faecalis strain SF28073 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |