Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 116137 |
| Name | oriT_59731|p59731-1 |
| Organism | Staphylococcus aureus strain 59731 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_AP024739 (26615..26803 [+], 189 nt) |
| oriT length | 189 nt |
| IRs (inverted repeats) | 149..155, 162..168 (CTATCAT..ATGATAG) 132..138, 142..148 (GTCTGGC..GCCAGAC) 71..76, 89..94 (AAAAAT..ATTTTT) 33..38, 45..50 (TGTCAC..GTGACA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 189 nt
>oriT_59731|p59731-1
TGTCACAAAAATAAGACAAAATCTACATGTAGTGTCACAAAACCGTGACAAACACAATATATAGTGTCACAAAAATGTGACACTAGGTATTTTTATGATATCACTATGAAATGTCCTAAAACCCTTGGAATGTCTGGCTTTGCCAGACCTATCATTGTACGATGATAGCAAATTCCCCTTATGCTCTTA
TGTCACAAAAATAAGACAAAATCTACATGTAGTGTCACAAAACCGTGACAAACACAATATATAGTGTCACAAAAATGTGACACTAGGTATTTTTATGATATCACTATGAAATGTCCTAAAACCCTTGGAATGTCTGGCTTTGCCAGACCTATCATTGTACGATGATAGCAAATTCCCCTTATGCTCTTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file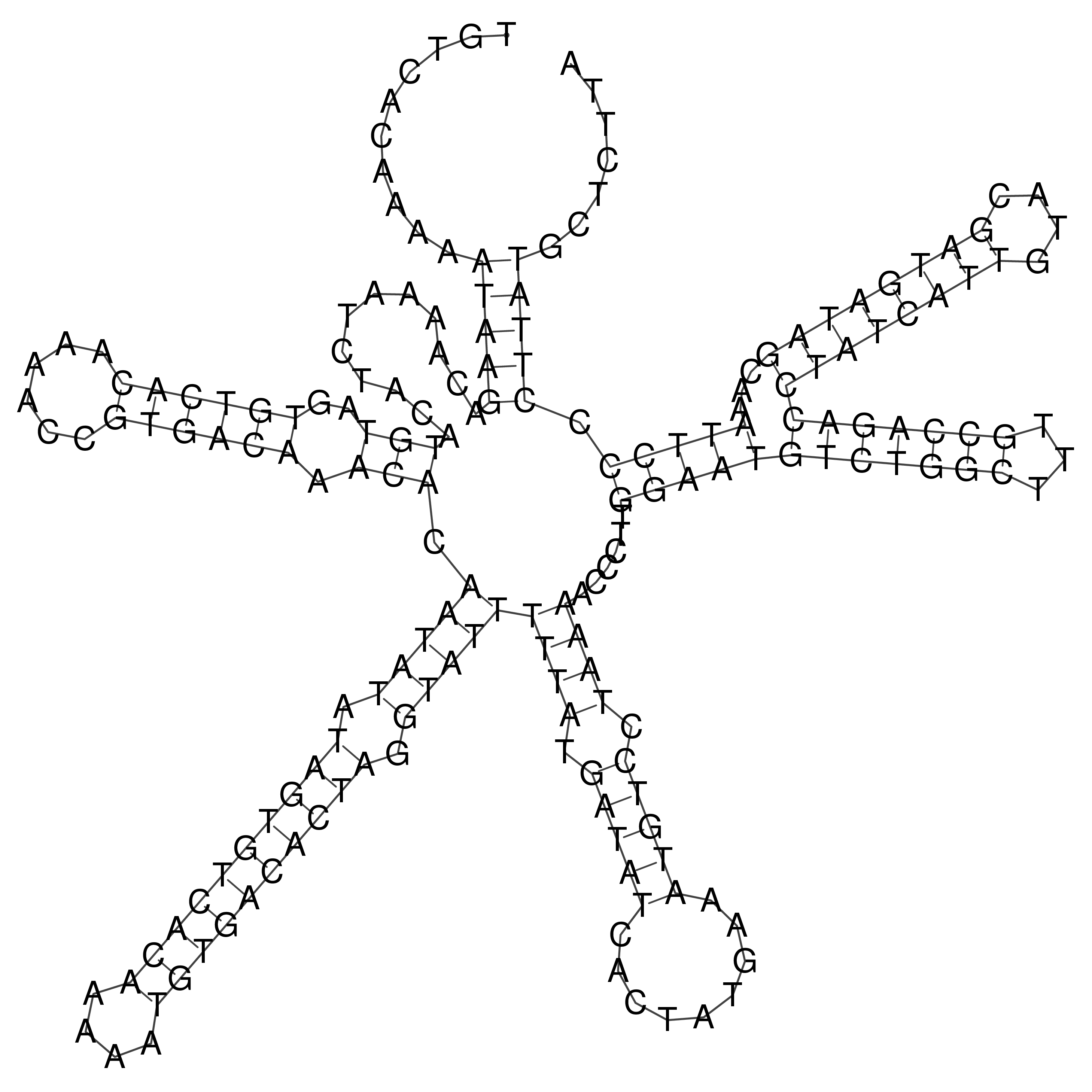
Relaxase
| ID | 10320 | GenBank | WP_042742193 |
| Name | mobP2_OCX20_RS14380_59731|p59731-1 |
UniProt ID | A0A1B3ISX3 |
| Length | 382 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 382 a.a. Molecular weight: 45794.09 Da Isoelectric Point: 9.8763
>WP_042742193.1 MobP2 family relaxase [Staphylococcus aureus]
MPKIVTVSQFEQPNEFYEYSNYVNYMKRGEAQNNKDDFKYDVYAHYMFDEEKSQNMFNEHDNFMSKKAIN
KIKSDFSHAQKNKGMMWKDVISFDNSSLESVGIYDSKTHTLDEQKIKEATRKMMKRFEKKEGLEDNLVWS
GAIHYNTDNIHVHVAAVEKVIKRTRGKRKPATLKHMKSTFANELFDIKGERQKINEFIRERIVKGIRDES
EPCKDKAMKKQIKKVHKLLEDIPKNQWNYNNNILKYIRPEIDKITDIYIQNYHSKDFEVFKNDLKRQNNL
YKETYGENSNYGSYEKTKMKDLYARSGNTILKNIKELDQDLMKVKQQFTHSKLNVTITKLKIGKCLNQAL
YRTKYALRSDYDKEKNIQQYYQEFDKQSYRER
MPKIVTVSQFEQPNEFYEYSNYVNYMKRGEAQNNKDDFKYDVYAHYMFDEEKSQNMFNEHDNFMSKKAIN
KIKSDFSHAQKNKGMMWKDVISFDNSSLESVGIYDSKTHTLDEQKIKEATRKMMKRFEKKEGLEDNLVWS
GAIHYNTDNIHVHVAAVEKVIKRTRGKRKPATLKHMKSTFANELFDIKGERQKINEFIRERIVKGIRDES
EPCKDKAMKKQIKKVHKLLEDIPKNQWNYNNNILKYIRPEIDKITDIYIQNYHSKDFEVFKNDLKRQNNL
YKETYGENSNYGSYEKTKMKDLYARSGNTILKNIKELDQDLMKVKQQFTHSKLNVTITKLKIGKCLNQAL
YRTKYALRSDYDKEKNIQQYYQEFDKQSYRER
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A1B3ISX3 |
T4CP
| ID | 12067 | GenBank | WP_235854108 |
| Name | t4cp2_OCX20_RS14330_59731|p59731-1 |
UniProt ID | _ |
| Length | 857 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 857 a.a. Molecular weight: 98676.15 Da Isoelectric Point: 6.2583
>WP_235854108.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Staphylococcus]
MKQLNTKYTDYIFNFRSDAVLLYIPIWVLVFIFFGKKVYQFRQRFKSLNNNEKASGRFATRKEITQQYKA
IPQREHAFEGEGGLPVAMFGVYDFVTKTKQYLEQRKTAIREINEITDLTADEKLELRKDKLGKMNKEFAT
TFFSKREFTFIDESPTNNLIVGTSRSGKGEIVVLSMIENYSRSSEQPSLIVNDMKGELFSASYKTLENRG
YQVEIFNLDQPMESTMSFNPLQQVIDEYMKGDLGEAQEMTKQITYTLTNNEGVEDNEWNLMAGALINAMI
LALCEETLPKNPEKVTLYSVSNMLQTLSTKRFYKEIAIGNKVQLIEVSALDEYMERFPSHSPAKTQYATV
QAAPDKMRSSIIGTALKALQPFTTDTIAKLTSHTSFDINKLGFPSIIDGYATKDSTFELGVQVLRDGENG
IEYEEVEGIQIGVNEYGYWQYPFKTVLKVGDRIETIEKDENNQVITSYFYVEHIDDKGQVAFNQKGELDN
SDITIRKFNSFPKPIAVFMVVPDYNNANHGIASILVNQIVFEISKNSQLYTENQSTYRRVIFHLDEAGNM
PPIPNLSQKVNVSLSRGLRFNFFVQAFSQFKDKYGDAYDAIMDACQNKVYIMATQEQTLKDFSAMIGSQQ
ITVHSRSGEAGAIKSSITENQEERPLLRPDELARLQEGETVVVRNLKRQDKKRNKVMSYPIFNDGKYAMK
YRYQYLSDLMDTSQSIIHFRPMLKARCEHRHLQLEATLVDWDKLIEEMQEGIDGQKEENKNINDINQYKK
SNEEGNVAVRVKRLETLKDKVGDAVFDKITRRFERYLRTYAEQGKNVNTITQKKLEELVLADSEVKEEEK
TKRIEMIKKLIKEAKTT
MKQLNTKYTDYIFNFRSDAVLLYIPIWVLVFIFFGKKVYQFRQRFKSLNNNEKASGRFATRKEITQQYKA
IPQREHAFEGEGGLPVAMFGVYDFVTKTKQYLEQRKTAIREINEITDLTADEKLELRKDKLGKMNKEFAT
TFFSKREFTFIDESPTNNLIVGTSRSGKGEIVVLSMIENYSRSSEQPSLIVNDMKGELFSASYKTLENRG
YQVEIFNLDQPMESTMSFNPLQQVIDEYMKGDLGEAQEMTKQITYTLTNNEGVEDNEWNLMAGALINAMI
LALCEETLPKNPEKVTLYSVSNMLQTLSTKRFYKEIAIGNKVQLIEVSALDEYMERFPSHSPAKTQYATV
QAAPDKMRSSIIGTALKALQPFTTDTIAKLTSHTSFDINKLGFPSIIDGYATKDSTFELGVQVLRDGENG
IEYEEVEGIQIGVNEYGYWQYPFKTVLKVGDRIETIEKDENNQVITSYFYVEHIDDKGQVAFNQKGELDN
SDITIRKFNSFPKPIAVFMVVPDYNNANHGIASILVNQIVFEISKNSQLYTENQSTYRRVIFHLDEAGNM
PPIPNLSQKVNVSLSRGLRFNFFVQAFSQFKDKYGDAYDAIMDACQNKVYIMATQEQTLKDFSAMIGSQQ
ITVHSRSGEAGAIKSSITENQEERPLLRPDELARLQEGETVVVRNLKRQDKKRNKVMSYPIFNDGKYAMK
YRYQYLSDLMDTSQSIIHFRPMLKARCEHRHLQLEATLVDWDKLIEEMQEGIDGQKEENKNINDINQYKK
SNEEGNVAVRVKRLETLKDKVGDAVFDKITRRFERYLRTYAEQGKNVNTITQKKLEELVLADSEVKEEEK
TKRIEMIKKLIKEAKTT
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 16570 | GenBank | NZ_AP024739 |
| Plasmid name | 59731|p59731-1 | Incompatibility group | - |
| Plasmid size | 40197 bp | Coordinate of oriT [Strand] | 26615..26803 [+] |
| Host baterium | Staphylococcus aureus strain 59731 |
Cargo genes
| Drug resistance gene | aac(6')-aph(2'') |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21, AcrIIA14 |