Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 116061 |
| Name | oriT_pSALNBL75 |
| Organism | Staphylococcus aureus strain B2-7A |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP042047 (19071..19246 [-], 176 nt) |
| oriT length | 176 nt |
| IRs (inverted repeats) | 154..159, 167..172 (AGCATA..TATGCT) 137..144, 148..155 (CTATCATT..AATGATAG) 120..126, 130..136 (GTCTGGC..GCCAGAC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 176 nt
>oriT_pSALNBL75
TGAGACATATACTATATATAGTGTCACAAAAGTGAGACAACCACTATATATAGTGTCACAAAGGTGAGACAAAAGGGGATTTAAGATATCATTTAAAAATTACCTTGAATTGTTGGAATGTCTGGCTTCGCCAGACCTATCATTATAAATGATAGCATATTCTCCTTATGCTCTTA
TGAGACATATACTATATATAGTGTCACAAAAGTGAGACAACCACTATATATAGTGTCACAAAGGTGAGACAAAAGGGGATTTAAGATATCATTTAAAAATTACCTTGAATTGTTGGAATGTCTGGCTTCGCCAGACCTATCATTATAAATGATAGCATATTCTCCTTATGCTCTTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file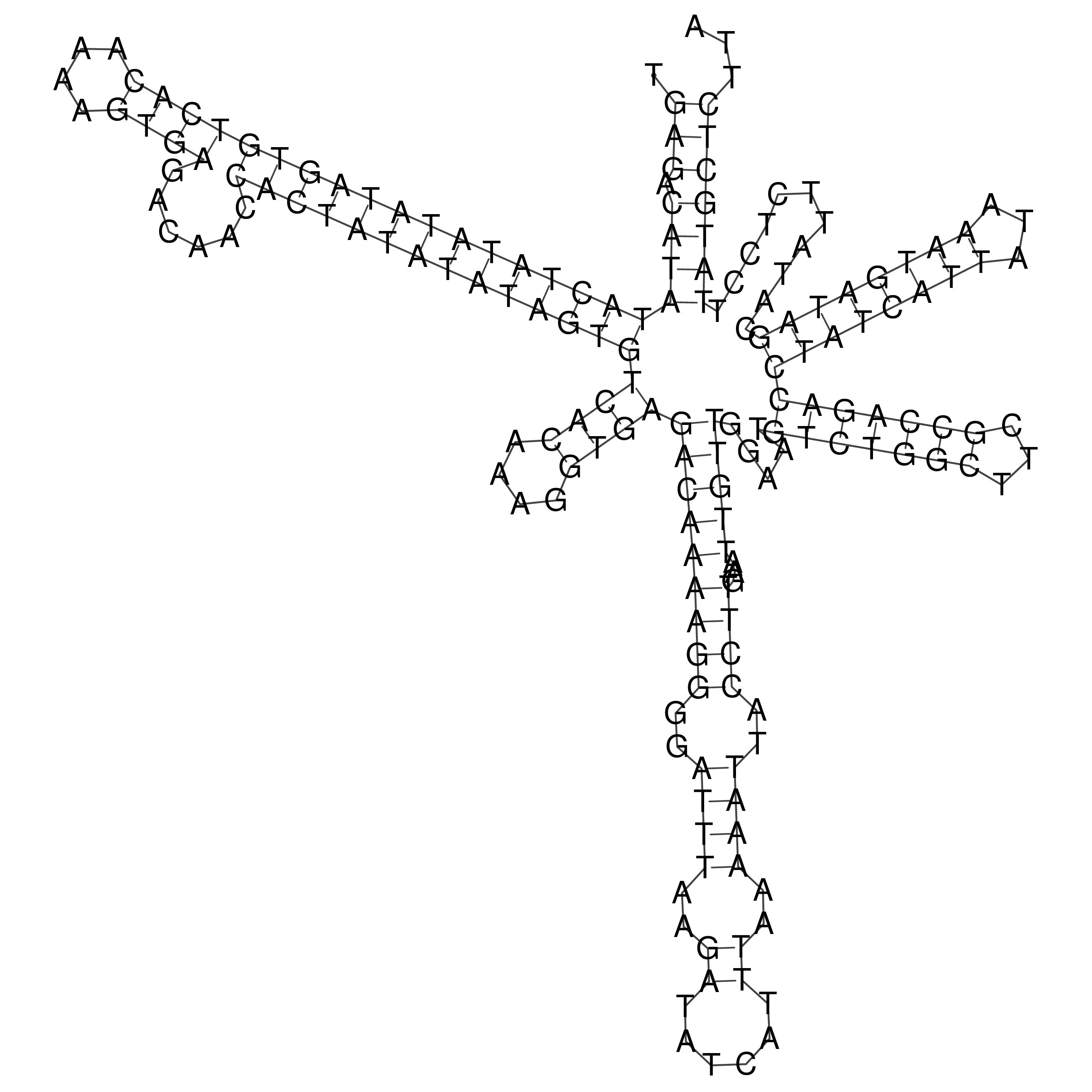
Relaxase
| ID | 10271 | GenBank | WP_145389854 |
| Name | mobP2_FP477_RS14135_pSALNBL75 |
UniProt ID | _ |
| Length | 391 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 391 a.a. Molecular weight: 47186.01 Da Isoelectric Point: 9.2044
>WP_145389854.1 MobP2 family relaxase [Staphylococcus aureus]
MPKIVTMSEFEQSKNKTKFSKYVDYVDRDEAKKNEHKEFQYDVYTHYMFDEQKSNSMFNDNSNYMSEDEI
KYTKSKFNEAQKNNGIMWKDVISFDNESLKEAGIFDPECKYLDEQKMRQATRKAMQKFEKKEGLENNLVW
SSSIHYNTDNIHVHVAAVEKDVTRERGKRKYSTIKEMKSSFANELFDMSGERTKINNFIRDRIVKGIKEQ
NEPDYDKQMKLQLKKIHEEIKDIPKKEWQYNNNIMKQARPEIDKFTLMYIKNNHGEEFDEFKQDLKKQSK
LYKETYGDKSDYKKYEETKMNDLYSRAGNTVLKQIKSFNEEYNNIKFKEINKQDNKKPLNINPKVNNKMQ
IGYALNSTLYNTQRALRNDFQKERNINQYHYEFDKSYEKEE
MPKIVTMSEFEQSKNKTKFSKYVDYVDRDEAKKNEHKEFQYDVYTHYMFDEQKSNSMFNDNSNYMSEDEI
KYTKSKFNEAQKNNGIMWKDVISFDNESLKEAGIFDPECKYLDEQKMRQATRKAMQKFEKKEGLENNLVW
SSSIHYNTDNIHVHVAAVEKDVTRERGKRKYSTIKEMKSSFANELFDMSGERTKINNFIRDRIVKGIKEQ
NEPDYDKQMKLQLKKIHEEIKDIPKKEWQYNNNIMKQARPEIDKFTLMYIKNNHGEEFDEFKQDLKKQSK
LYKETYGDKSDYKKYEETKMNDLYSRAGNTVLKQIKSFNEEYNNIKFKEINKQDNKKPLNINPKVNNKMQ
IGYALNSTLYNTQRALRNDFQKERNINQYHYEFDKSYEKEE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 12005 | GenBank | WP_261374895 |
| Name | t4cp2_FP477_RS14185_pSALNBL75 |
UniProt ID | _ |
| Length | 941 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 941 a.a. Molecular weight: 108637.71 Da Isoelectric Point: 8.3862
>WP_261374895.1 VirD4-like conjugal transfer protein, CD1115 family [Staphylococcus aureus]
MIPFRSKHNHGYHRKKASFSQNKHLLAHPIGLLALLVVGFIFIMLAANFFINMLNNLIFAGGDIFDNMKK
INNEWTNYFFKVRFDLALFYIPFFVLLTVLWGKKVYQIRQKFKSLDNQEKASGRFSTKKEIKQQYKAIAQ
REKQFEGEGGLPVAMFGVYDFATKTKQYLEQRKTAKTEINSMPDLTQDEKYNMKKDKLGQINKEFVSTFF
LKREFTFIDESPTNNIFVGTSRSGKGEVFVLSMIENYSRSSHQPSLVVGDMKGELYSASNETLENRNYHT
EVFNLDQPMESTMSFNLLQFVIEEYQKGDLGEAQEMTKQITHTLTNNEGVEDNEWNLMSAALINAMILAL
CEEALPEHPEKVTLYSVSNMLQTLSVKKFYKQIQIGNQVKVIETSALDEYMERFPSHSPAKTQYATVEAA
PDKMRSSIIGTALKALQPFTTDTIAKLTSHSSLDLNKLGFPSIIDGFSEPDSKFELNVQVKRETNIGTKY
DEVEGINIGVNEYGYWQYPFKTVLNIDDRIEIVETDEFNEEVKYYFYVTSIDKKGQVTFEAKGDIYSSNV
TIRKFNSFAKPIAIFMVVPDYNSANHGIASILVNQIVFELSKNSQLYTEKQSTHRRVIFHLDEAGNMPQI
PNLSQKVNVSLSRGLRFNFFVQAFSQIKDTYGDAYDAIMDACQNKVFIMATQEQTLKDFSDMIGSQQITV
RSRSGETDSLKSSITENQEERPLLRADELAHLQEGETVVVRNLKRQDNKRRKVMSYPIFNSGKYAMKYRY
QYLADLMDTGKSIIHFRPLLKERCEHRHLQLETTLVDWEKIIENMQAGIENDSNNDIDDLTQSLNKSMGE
KANANNPEVYKEENNKKPVTRQKKVLTLKAQLSAEEFDRLTRRFTKTIRMYEKDGNNVDRLTLYKLKQYL
KLRVRNNDITEEQKNKSINYVKQLIEEAKAK
MIPFRSKHNHGYHRKKASFSQNKHLLAHPIGLLALLVVGFIFIMLAANFFINMLNNLIFAGGDIFDNMKK
INNEWTNYFFKVRFDLALFYIPFFVLLTVLWGKKVYQIRQKFKSLDNQEKASGRFSTKKEIKQQYKAIAQ
REKQFEGEGGLPVAMFGVYDFATKTKQYLEQRKTAKTEINSMPDLTQDEKYNMKKDKLGQINKEFVSTFF
LKREFTFIDESPTNNIFVGTSRSGKGEVFVLSMIENYSRSSHQPSLVVGDMKGELYSASNETLENRNYHT
EVFNLDQPMESTMSFNLLQFVIEEYQKGDLGEAQEMTKQITHTLTNNEGVEDNEWNLMSAALINAMILAL
CEEALPEHPEKVTLYSVSNMLQTLSVKKFYKQIQIGNQVKVIETSALDEYMERFPSHSPAKTQYATVEAA
PDKMRSSIIGTALKALQPFTTDTIAKLTSHSSLDLNKLGFPSIIDGFSEPDSKFELNVQVKRETNIGTKY
DEVEGINIGVNEYGYWQYPFKTVLNIDDRIEIVETDEFNEEVKYYFYVTSIDKKGQVTFEAKGDIYSSNV
TIRKFNSFAKPIAIFMVVPDYNSANHGIASILVNQIVFELSKNSQLYTEKQSTHRRVIFHLDEAGNMPQI
PNLSQKVNVSLSRGLRFNFFVQAFSQIKDTYGDAYDAIMDACQNKVFIMATQEQTLKDFSDMIGSQQITV
RSRSGETDSLKSSITENQEERPLLRADELAHLQEGETVVVRNLKRQDNKRRKVMSYPIFNSGKYAMKYRY
QYLADLMDTGKSIIHFRPLLKERCEHRHLQLETTLVDWEKIIENMQAGIENDSNNDIDDLTQSLNKSMGE
KANANNPEVYKEENNKKPVTRQKKVLTLKAQLSAEEFDRLTRRFTKTIRMYEKDGNNVDRLTLYKLKQYL
KLRVRNNDITEEQKNKSINYVKQLIEEAKAK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 16494 | GenBank | NZ_CP042047 |
| Plasmid name | pSALNBL75 | Incompatibility group | - |
| Plasmid size | 75938 bp | Coordinate of oriT [Strand] | 19071..19246 [-] |
| Host baterium | Staphylococcus aureus strain B2-7A |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |