Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 116014 |
| Name | oriT_NCDC 599-52|p1 |
| Organism | Shigella dysenteriae strain NCDC 599-52 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP119452 (14110..14233 [+], 124 nt) |
| oriT length | 124 nt |
| IRs (inverted repeats) | 92..99, 113..120 (ATAATGTA..TACATTAT) 90..95, 107..112 (AAATAA..TTATTT) 39..46, 49..56 (GCAAAAAC..GTTTTTGC) 3..10, 15..22 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 124 nt
GGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTGCTATTTGAATCATTAACTTATGTTTTAAATAATGTATTTTAATTTATTTTACATTATAAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file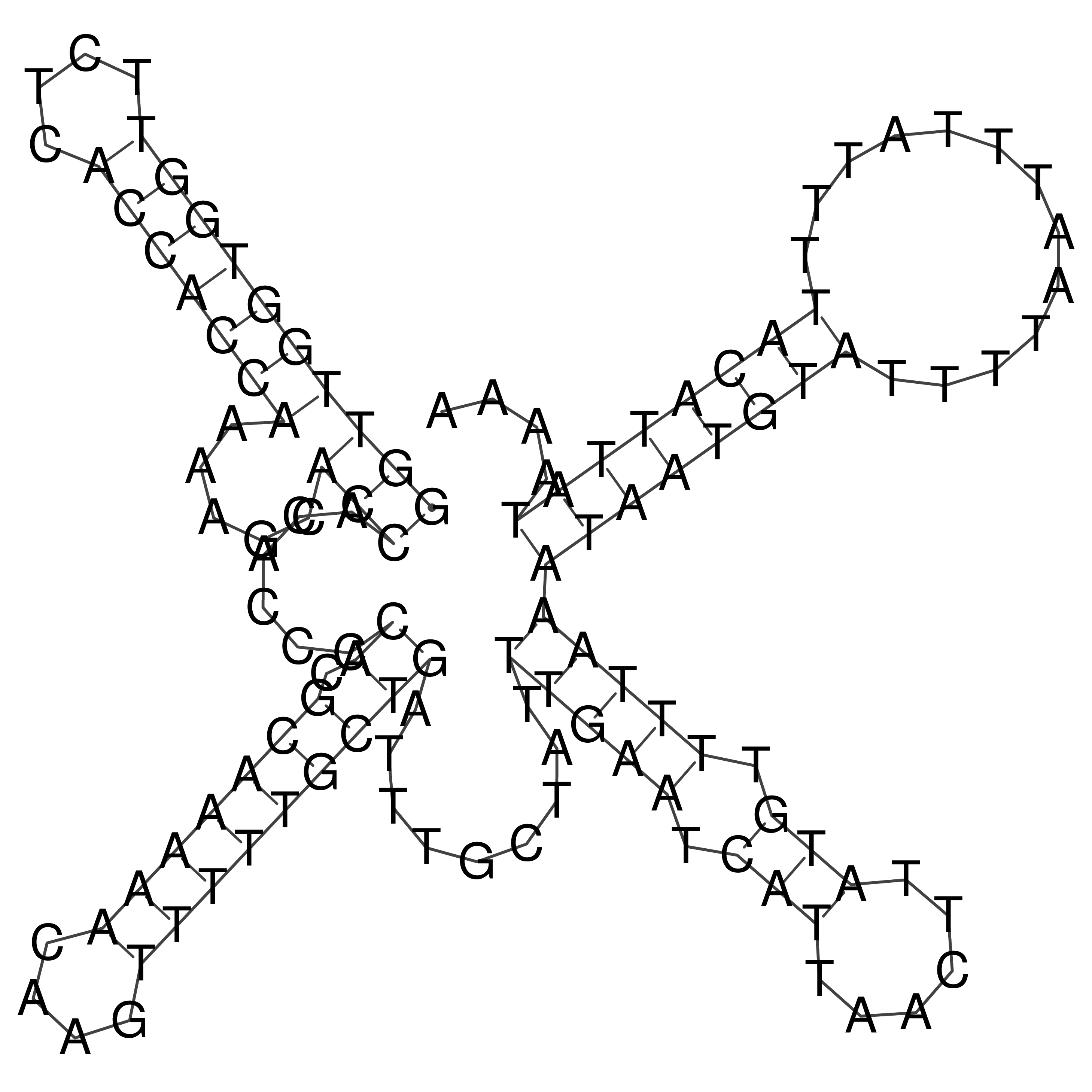
Relaxase
| ID | 10237 | GenBank | WP_315700217 |
| Name | traI_PWE30_RS25665_NCDC 599-52|p1 |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 191829.64 Da Isoelectric Point: 5.8498
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRIMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQTIREAVGEDASLKSRDVAALDTRKSKQHVDPEIKMAEWMQT
LKETGFDIRAYRDAADQRAETRTLTPGPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTIHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLQEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFGASVKAGEESVAQVSGVREQAMLTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKIPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDAMTVVVPGRAEPATLPVVDSPFTALKL
ENGWVETPGHSVSDSATVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETSLETAISLQKAGLHTPAQQAIHLALPVLESKNLAFSMVDLLTEAKSFAAEGTGFTELGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMETLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPASERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVVIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMQKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWEKNPDALALVDSVYHRIAGISRDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDKIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQEQAEQHIDLAYAI
TVHGAQGASETFAIALEGTEGNRKLMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINKTVQKGTAHDVL
EPGADREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGRSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPRAITGGRVWGDIPDNSLQPGAGNGEPVTAEVLAQRQAEEAIRCETERRADEIVRKMAENKPDLP
DGRTEQAVREIVGQERDRTVTSEGEAVLPESVLREPQRVREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 6026 | GenBank | WP_001151564 |
| Name | WP_001151564_NCDC 599-52|p1 |
UniProt ID | A0AA36PD43 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14446.39 Da Isoelectric Point: 4.7197
MAKVQAYVSDEIVYKINKIVERRRAEGAKSTDVSFSSISTMLLELGLRVYEAQMERKESAFNQAEFNKVL
LECAVKTQSTVAKILGIESLSPHVSGNPKFEYANMVEDIRDKVSSEMERFFPENDEE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 6027 | GenBank | WP_001254388 |
| Name | WP_001254388_NCDC 599-52|p1 |
UniProt ID | W9A0Z2 |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 9072.20 Da Isoelectric Point: 10.2071
MRRRNARGGISRTVSVYLDEDTNNRLIRAKDRSGRSKTIEVQIRLRDHLKRFPDFYNEEIFREVIEENES
TFKEL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | W9A0Z2 |
T4CP
| ID | 11961 | GenBank | WP_032250496 |
| Name | traC_PWE30_RS25565_NCDC 599-52|p1 |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 99304.97 Da Isoelectric Point: 6.0793
MSNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAI
PINGANESVVEALDHMLRTKLPRGVPFCIHLMSSQLVGDRIEYGLREFSWSGEQAVRFNAITRAYYMRAA
ATQFPLPEGMNLPLTLRHYRVFFSYCSPSKKKSRADILEMENLVKIIRASLQGASITTQTVDAQAFIDIV
GEMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAF
LWNMADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWG
ELRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGK
GLFKQLKEAGVIQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSG
AGKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLS
VMASPNGNLDEVHEGLLLQAVRASWLAKENRARIDDVVDFLKNASDSEQYAGSPTIRSRLDEMIVLLDQY
TANGTYGQYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGW
RLLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKY
NQLYPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRREG
MSIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 11962 | GenBank | WP_128610773 |
| Name | traD_PWE30_RS25660_NCDC 599-52|p1 |
UniProt ID | _ |
| Length | 735 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 735 a.a. Molecular weight: 83653.31 Da Isoelectric Point: 5.0541
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLILWVKISWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLATVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTDNPKDVARMLKKDDKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
KRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGEPFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQTRPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPDVPEVVSGEDVTQAEQPQQPQQPQQ
PQQPQQPQQPQQPVSPAINDKKSDSGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAAYEAWQ
QENHPDTQQQMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 13538..41815
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| PWE30_RS25440 (PWE30_25385) | 8900..9154 | + | 255 | WP_032250339 | hypothetical protein | - |
| PWE30_RS25445 (PWE30_25390) | 9151..9393 | + | 243 | WP_000151456 | hypothetical protein | - |
| PWE30_RS25450 (PWE30_25395) | 9964..10257 | + | 294 | WP_000673428 | hypothetical protein | - |
| PWE30_RS25455 (PWE30_25400) | 10562..10849 | + | 288 | WP_100037732 | hypothetical protein | - |
| PWE30_RS25460 (PWE30_25405) | 11277..11510 | + | 234 | WP_000093744 | hypothetical protein | - |
| PWE30_RS25465 (PWE30_25410) | 11586..11888 | + | 303 | WP_000379301 | hypothetical protein | - |
| PWE30_RS25470 (PWE30_25415) | 12160..12309 | + | 150 | WP_000903193 | hypothetical protein | - |
| PWE30_RS25475 (PWE30_25420) | 12420..13241 | + | 822 | WP_073975216 | DUF932 domain-containing protein | - |
| PWE30_RS25480 (PWE30_25425) | 13538..14185 | - | 648 | WP_000614287 | transglycosylase SLT domain-containing protein | virB1 |
| PWE30_RS25485 (PWE30_25430) | 14471..14854 | + | 384 | WP_001151564 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| PWE30_RS25490 (PWE30_25435) | 15048..15734 | + | 687 | WP_000332487 | PAS domain-containing protein | - |
| PWE30_RS25495 (PWE30_25440) | 15828..16055 | + | 228 | WP_001254388 | conjugal transfer relaxosome protein TraY | - |
| PWE30_RS25500 (PWE30_25445) | 16089..16448 | + | 360 | WP_019842162 | type IV conjugative transfer system pilin TraA | - |
| PWE30_RS25505 (PWE30_25450) | 16463..16774 | + | 312 | WP_032250594 | type IV conjugative transfer system protein TraL | traL |
| PWE30_RS25510 (PWE30_25455) | 16796..17362 | + | 567 | WP_000399805 | type IV conjugative transfer system protein TraE | traE |
| PWE30_RS25515 (PWE30_25460) | 17349..18077 | + | 729 | WP_001230787 | type-F conjugative transfer system secretin TraK | traK |
| PWE30_RS25520 (PWE30_25465) | 18077..19504 | + | 1428 | WP_315700229 | F-type conjugal transfer pilus assembly protein TraB | traB |
| PWE30_RS25525 (PWE30_25470) | 19494..20084 | + | 591 | WP_000002782 | conjugal transfer pilus-stabilizing protein TraP | - |
| PWE30_RS25530 (PWE30_25475) | 20071..20391 | + | 321 | WP_001057306 | conjugal transfer protein TrbD | virb4 |
| PWE30_RS25535 (PWE30_25480) | 20384..20635 | + | 252 | WP_001061854 | conjugal transfer protein TrbG | - |
| PWE30_RS25540 (PWE30_25485) | 20632..21147 | + | 516 | WP_032324751 | type IV conjugative transfer system lipoprotein TraV | traV |
| PWE30_RS25545 (PWE30_25490) | 21282..21503 | + | 222 | WP_001278695 | conjugal transfer protein TraR | - |
| PWE30_RS25550 (PWE30_25495) | 21496..21972 | + | 477 | WP_032250499 | hypothetical protein | - |
| PWE30_RS25555 (PWE30_25500) | 22052..22270 | + | 219 | WP_032250498 | hypothetical protein | - |
| PWE30_RS25560 (PWE30_25505) | 22298..22645 | + | 348 | WP_315700231 | hypothetical protein | - |
| PWE30_RS25565 (PWE30_25510) | 22771..25401 | + | 2631 | WP_032250496 | type IV secretion system protein TraC | virb4 |
| PWE30_RS25570 (PWE30_25515) | 25398..25784 | + | 387 | WP_000214082 | type-F conjugative transfer system protein TrbI | - |
| PWE30_RS25575 (PWE30_25520) | 25781..26413 | + | 633 | WP_032250504 | type-F conjugative transfer system protein TraW | traW |
| PWE30_RS25580 (PWE30_25525) | 26410..27402 | + | 993 | WP_000830838 | conjugal transfer pilus assembly protein TraU | traU |
| PWE30_RS25585 (PWE30_25530) | 27411..28049 | + | 639 | WP_001744737 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| PWE30_RS25590 (PWE30_25535) | 28046..29896 | + | 1851 | WP_000821864 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| PWE30_RS25595 (PWE30_25540) | 29923..30180 | + | 258 | WP_000864329 | conjugal transfer protein TrbE | - |
| PWE30_RS25600 (PWE30_25545) | 30173..30916 | + | 744 | WP_001030371 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| PWE30_RS25605 (PWE30_25550) | 30930..31271 | + | 342 | WP_000556795 | conjugal transfer protein TrbA | - |
| PWE30_RS25610 (PWE30_25555) | 31252..31587 | - | 336 | WP_000736414 | hypothetical protein | - |
| PWE30_RS25615 (PWE30_25560) | 31668..31952 | + | 285 | WP_021566306 | type-F conjugative transfer system pilin chaperone TraQ | - |
| PWE30_RS25620 (PWE30_25565) | 31939..32484 | + | 546 | WP_000059830 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| PWE30_RS25625 (PWE30_25570) | 32414..32761 | + | 348 | WP_072006150 | P-type conjugative transfer protein TrbJ | - |
| PWE30_RS25630 (PWE30_25575) | 32742..33134 | + | 393 | WP_032250489 | F-type conjugal transfer protein TrbF | - |
| PWE30_RS25635 (PWE30_25580) | 33121..34494 | + | 1374 | WP_021501421 | conjugal transfer pilus assembly protein TraH | traH |
| PWE30_RS25640 (PWE30_25585) | 34491..37307 | + | 2817 | WP_126739281 | conjugal transfer mating-pair stabilization protein TraG | traG |
| PWE30_RS25645 (PWE30_25590) | 37340..37861 | + | 522 | WP_000632668 | conjugal transfer entry exclusion protein TraS | - |
| PWE30_RS25650 (PWE30_25595) | 37886..38617 | + | 732 | WP_000850429 | conjugal transfer complement resistance protein TraT | - |
| PWE30_RS25655 (PWE30_25600) | 38820..39557 | + | 738 | WP_062877098 | hypothetical protein | - |
| PWE30_RS25660 (PWE30_25605) | 39608..41815 | + | 2208 | WP_128610773 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 16447 | GenBank | NZ_CP119452 |
| Plasmid name | NCDC 599-52|p1 | Incompatibility group | - |
| Plasmid size | 51197 bp | Coordinate of oriT [Strand] | 14110..14233 [+] |
| Host baterium | Shigella dysenteriae strain NCDC 599-52 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |