Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 115875 |
| Name | oriT_3.2.17|unnamed |
| Organism | Enterobacter cancerogenus strain 3.2.17 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP134405 (74637..74749 [+], 113 nt) |
| oriT length | 113 nt |
| IRs (inverted repeats) | 93..99, 103..109 (TAATTTA..TAAATTA) 90..95, 97..102 (AAATAA..TTATTT) 29..34, 41..46 (GCAAAA..TTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 113 nt
>oriT_3.2.17|unnamed
TTCTCACCACAAAAGCACCACACCCCACGCAAAACGTACTTTTTGCTGATTGGCTATTCAGTTCAGTAAGTTAGCGTTTGAATAATGTGAAATAATTTATTTTAAATTACTAA
TTCTCACCACAAAAGCACCACACCCCACGCAAAACGTACTTTTTGCTGATTGGCTATTCAGTTCAGTAAGTTAGCGTTTGAATAATGTGAAATAATTTATTTTAAATTACTAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file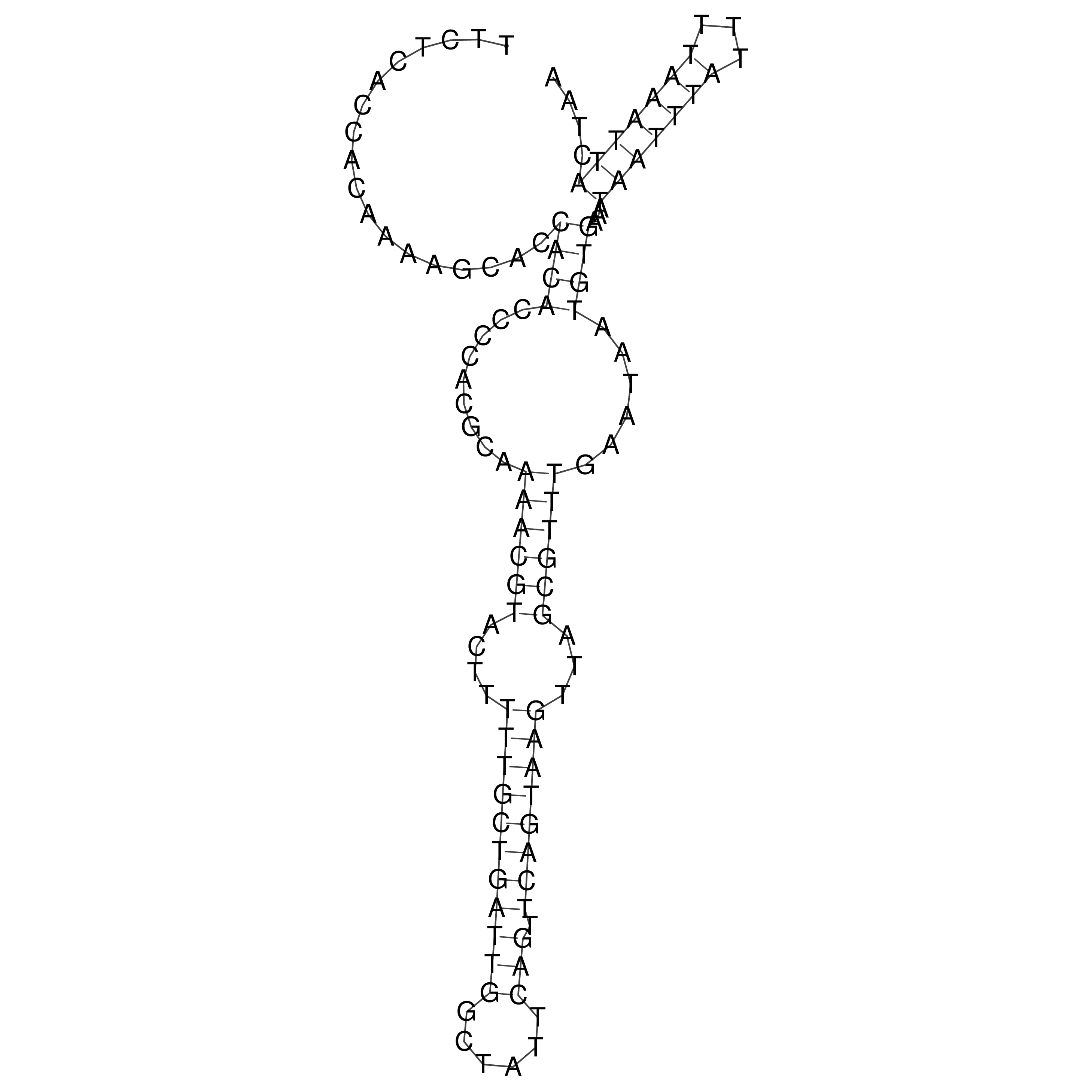
Relaxase
| ID | 10165 | GenBank | WP_313830629 |
| Name | traI_RIN64_RS23120_3.2.17|unnamed |
UniProt ID | _ |
| Length | 1747 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1747 a.a. Molecular weight: 189258.36 Da Isoelectric Point: 6.3948
>WP_313830629.1 conjugative transfer relaxase/helicase TraI [Enterobacter cancerogenus]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGEGAKALGLSGSVDVKTFTRVLEGKLPDGSDLSRAQ
DGTNKHRPGYDLTFSAPKSISVMAMLGGDTRLIDAHNRAVDTAIKQIEALASTRVMTEGKSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHDGTWRTLSSDKIGKTGFIENVYANQIAFGRLYRAALKDDVTA
MGYETETVGKHGMWELKGVPTEPYSSRSKAIREAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMAT
LKETGFDIQAYREAADQRARGGDIPVTTPEPVNVDTAVGQSIAMLSDRRARFTYSELLATTLGQLPARPG
MIEQAREGIDAAIKNEQLIPLDKEKGLFTSNVHVLDELSVSALSLELQQQGRVDVFPDKSVPRSRAFGDA
VSVLAQDRSPVAIISGQGGAAGQRERVAELAMMAREQGRDVQIMAADRRSAANLARDERLSGELITDRRG
LTEGMTFIPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTRVSVISEPDRRERYDRLAADFAKSIRAGEEAVVQISGAREQAALAATVRDALKGEKVLGERDISIT
TLEPVWLDGRHRQVRDHYREGMVMERWNAEERSRERFVIDRVTSRNNSLTLRNAQGETLVTRLTELDSSW
SLYRTGTLQVTEGDRLAVLGQTQGARLKGGDSVTVTGIDEKGIHVSLPGRKTQAVLPAGDSPFTAPKIGQ
GWVESPGRSVSDSATVYASLTQREMDNATLNGLARSGSEIRLYSAQTVQKTEEKLSRQSAWTVVTAQVKE
AAGKDSLGEALAHQKAALHTPEQQAIHLAIPSLEGNGLAFTRPQLMAAAKDFAQDKLSLAAIEDEIARQT
RSGALISVPVSQGNGLQQLVSRQSYNAEKSIIRHVLEGKDAITPLMGKVPDAQLAGLTDGQQNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEEQRPRVIGVAPTHRAVGEMRDAGVPESQTLAAFIHDTQ
QQLRGGERPDFSNVLFLIDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DTVVMQEIVRQTPALRPAVYSLIERNIGSALTTLESVAPQQVPRRPDAWQPDGSVMEFSREQEKAIADAI
KAGDLAPGGQPATLLEAIVKDYTGRTPQAQAQTIVITALNADRRQLNAMIHDARREAGELGEKEAIVPVL
TPANIRDGELRRMDTWQTHASSKVLLDNTYYRIDALDKDAHLVTLKDEQGNARSLSPAQAATEGVTLYRQ
DTIAVSPGDRMRFSKSDNDRGFVANSVWTVSDIKGDSVTLTDGKQTRTVRPGVEPAEQHIDLAYAVTVYG
AQGASEPFSISLQGTDGGRKQMVNFESAYVALSRMKQHAQVYTDNREKWVAAMEKSQAKSTAHDIVEPRN
DCAVANAARLTASAKGLGEVPAGRAALRQAGLRPETTLAKFISPGRKYPQPHVALPAFDRNGRQAGVWLS
TLTDGDGRLQGLSGERRVMGSGDAAFAGLQASRNGESLMAGDMTEGVRLARENPQSGVVVRIGDAEGRPW
NPGAITGGRVWADVVPDGTGTQHGEKIPPEVLAQQALEAQQRRELEKRAEDAVRELARGGEKSAEPADAV
RELARELGEGKNRERSTDVTLPESPDVRARDEAVSRVAHENVQRDRLQQMERDTVRVLEREKTQGGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGEGAKALGLSGSVDVKTFTRVLEGKLPDGSDLSRAQ
DGTNKHRPGYDLTFSAPKSISVMAMLGGDTRLIDAHNRAVDTAIKQIEALASTRVMTEGKSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHDGTWRTLSSDKIGKTGFIENVYANQIAFGRLYRAALKDDVTA
MGYETETVGKHGMWELKGVPTEPYSSRSKAIREAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMAT
LKETGFDIQAYREAADQRARGGDIPVTTPEPVNVDTAVGQSIAMLSDRRARFTYSELLATTLGQLPARPG
MIEQAREGIDAAIKNEQLIPLDKEKGLFTSNVHVLDELSVSALSLELQQQGRVDVFPDKSVPRSRAFGDA
VSVLAQDRSPVAIISGQGGAAGQRERVAELAMMAREQGRDVQIMAADRRSAANLARDERLSGELITDRRG
LTEGMTFIPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTRVSVISEPDRRERYDRLAADFAKSIRAGEEAVVQISGAREQAALAATVRDALKGEKVLGERDISIT
TLEPVWLDGRHRQVRDHYREGMVMERWNAEERSRERFVIDRVTSRNNSLTLRNAQGETLVTRLTELDSSW
SLYRTGTLQVTEGDRLAVLGQTQGARLKGGDSVTVTGIDEKGIHVSLPGRKTQAVLPAGDSPFTAPKIGQ
GWVESPGRSVSDSATVYASLTQREMDNATLNGLARSGSEIRLYSAQTVQKTEEKLSRQSAWTVVTAQVKE
AAGKDSLGEALAHQKAALHTPEQQAIHLAIPSLEGNGLAFTRPQLMAAAKDFAQDKLSLAAIEDEIARQT
RSGALISVPVSQGNGLQQLVSRQSYNAEKSIIRHVLEGKDAITPLMGKVPDAQLAGLTDGQQNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEEQRPRVIGVAPTHRAVGEMRDAGVPESQTLAAFIHDTQ
QQLRGGERPDFSNVLFLIDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DTVVMQEIVRQTPALRPAVYSLIERNIGSALTTLESVAPQQVPRRPDAWQPDGSVMEFSREQEKAIADAI
KAGDLAPGGQPATLLEAIVKDYTGRTPQAQAQTIVITALNADRRQLNAMIHDARREAGELGEKEAIVPVL
TPANIRDGELRRMDTWQTHASSKVLLDNTYYRIDALDKDAHLVTLKDEQGNARSLSPAQAATEGVTLYRQ
DTIAVSPGDRMRFSKSDNDRGFVANSVWTVSDIKGDSVTLTDGKQTRTVRPGVEPAEQHIDLAYAVTVYG
AQGASEPFSISLQGTDGGRKQMVNFESAYVALSRMKQHAQVYTDNREKWVAAMEKSQAKSTAHDIVEPRN
DCAVANAARLTASAKGLGEVPAGRAALRQAGLRPETTLAKFISPGRKYPQPHVALPAFDRNGRQAGVWLS
TLTDGDGRLQGLSGERRVMGSGDAAFAGLQASRNGESLMAGDMTEGVRLARENPQSGVVVRIGDAEGRPW
NPGAITGGRVWADVVPDGTGTQHGEKIPPEVLAQQALEAQQRRELEKRAEDAVRELARGGEKSAEPADAV
RELARELGEGKNRERSTDVTLPESPDVRARDEAVSRVAHENVQRDRLQQMERDTVRVLEREKTQGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 11875 | GenBank | WP_313830627 |
| Name | traD_RIN64_RS23115_3.2.17|unnamed |
UniProt ID | _ |
| Length | 746 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 746 a.a. Molecular weight: 84085.92 Da Isoelectric Point: 5.0963
>WP_313830627.1 type IV conjugative transfer system coupling protein TraD [Enterobacter cancerogenus]
MSFNAKDMTQGGQIASMRIRMFSQIANIILYCLVILFGIIVALTLAVKLSWQTFVNGMAWYWCQTLAPMR
DIFRSQPVYTIRYYGKELKYNAQQLLQDKYVIWCADQVLTAFILAAIIAAVVCVIAFFVTTWILGHQGKQ
QSEDEVTGGRQLTDDPREVARMLKRDNLASDIKIGDLPLIKNSEIQNFCLHGTVGAGKSEVIRRLMNYAR
KRGDMVIVYDRSCEFVKSYYDPAIDKILNPMDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLQNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGEPFTIRDWMRGVREDGRNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDIQTLPDLSCYVTLPGPYPAVKMAL
KYQERPKVAPEFIQRRIDDKAEQRLNAALAAREAESRSVAMLFEPDTEVVAPVAGPENAVQPQQPQQPQQ
PQQPQQPQQPQQPQQPQQPQQPQPAPVITPRPAPVAASTSVAGTGGTESVLETKAEEAEQLPPGIDESGE
VVDMAAWEAWQAEQDELTPQERQRREEVNINVHRTHDDDLEPGGYI
MSFNAKDMTQGGQIASMRIRMFSQIANIILYCLVILFGIIVALTLAVKLSWQTFVNGMAWYWCQTLAPMR
DIFRSQPVYTIRYYGKELKYNAQQLLQDKYVIWCADQVLTAFILAAIIAAVVCVIAFFVTTWILGHQGKQ
QSEDEVTGGRQLTDDPREVARMLKRDNLASDIKIGDLPLIKNSEIQNFCLHGTVGAGKSEVIRRLMNYAR
KRGDMVIVYDRSCEFVKSYYDPAIDKILNPMDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLQNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGEPFTIRDWMRGVREDGRNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDIQTLPDLSCYVTLPGPYPAVKMAL
KYQERPKVAPEFIQRRIDDKAEQRLNAALAAREAESRSVAMLFEPDTEVVAPVAGPENAVQPQQPQQPQQ
PQQPQQPQQPQQPQQPQQPQQPQPAPVITPRPAPVAASTSVAGTGGTESVLETKAEEAEQLPPGIDESGE
VVDMAAWEAWQAEQDELTPQERQRREEVNINVHRTHDDDLEPGGYI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 74095..97075
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| RIN64_RS22960 (RIN64_22960) | 69216..69395 | + | 180 | Protein_79 | theronine dehydrogenase | - |
| RIN64_RS22965 (RIN64_22965) | 69568..69717 | + | 150 | Protein_80 | DUF5431 family protein | - |
| RIN64_RS22970 (RIN64_22970) | 69665..69784 | + | 120 | WP_272929840 | type I toxin-antitoxin system Hok family toxin | - |
| RIN64_RS22975 (RIN64_22975) | 70487..70876 | + | 390 | WP_313830589 | ammonia monooxygenase | - |
| RIN64_RS22980 (RIN64_22980) | 70938..71219 | + | 282 | WP_237658073 | hypothetical protein | - |
| RIN64_RS22985 (RIN64_22985) | 71266..71622 | + | 357 | WP_063449773 | hypothetical protein | - |
| RIN64_RS22990 (RIN64_22990) | 71708..71971 | + | 264 | WP_046622508 | hypothetical protein | - |
| RIN64_RS22995 (RIN64_22995) | 72017..72847 | + | 831 | WP_313830591 | N-6 DNA methylase | - |
| RIN64_RS23000 (RIN64_23000) | 73525..74055 | + | 531 | WP_313830593 | antirestriction protein | - |
| RIN64_RS23005 (RIN64_23005) | 74095..74568 | - | 474 | WP_313830595 | transglycosylase SLT domain-containing protein | virB1 |
| RIN64_RS23010 (RIN64_23010) | 74999..75394 | + | 396 | WP_313830597 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| RIN64_RS23015 (RIN64_23015) | 75578..76282 | + | 705 | WP_193037203 | LuxR C-terminal-related transcriptional regulator | - |
| RIN64_RS23020 (RIN64_23020) | 76361..76585 | + | 225 | WP_046622499 | TraY domain-containing protein | - |
| RIN64_RS23025 (RIN64_23025) | 76632..76982 | + | 351 | WP_202562312 | type IV conjugative transfer system pilin TraA | - |
| RIN64_RS23030 (RIN64_23030) | 77119..77424 | + | 306 | WP_063449767 | type IV conjugative transfer system protein TraL | traL |
| RIN64_RS23035 (RIN64_23035) | 77439..78005 | + | 567 | WP_046622494 | type IV conjugative transfer system protein TraE | traE |
| RIN64_RS23040 (RIN64_23040) | 77992..78735 | + | 744 | WP_313830604 | type-F conjugative transfer system secretin TraK | traK |
| RIN64_RS23045 (RIN64_23045) | 78722..80125 | + | 1404 | WP_313830606 | F-type conjugal transfer pilus assembly protein TraB | traB |
| RIN64_RS23050 (RIN64_23050) | 80146..80682 | + | 537 | WP_202562307 | type IV conjugative transfer system lipoprotein TraV | traV |
| RIN64_RS23055 (RIN64_23055) | 80989..83619 | + | 2631 | WP_313830609 | type IV secretion system protein TraC | virb4 |
| RIN64_RS23060 (RIN64_23060) | 83616..83963 | + | 348 | WP_313830611 | type-F conjugative transfer system protein TrbI | - |
| RIN64_RS23065 (RIN64_23065) | 83963..84610 | + | 648 | WP_313830613 | type-F conjugative transfer system protein TraW | traW |
| RIN64_RS23070 (RIN64_23070) | 84607..85593 | + | 987 | WP_063449758 | conjugal transfer pilus assembly protein TraU | traU |
| RIN64_RS23075 (RIN64_23075) | 85608..86231 | + | 624 | WP_313830616 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| RIN64_RS23080 (RIN64_23080) | 86228..88114 | + | 1887 | WP_313830618 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| RIN64_RS23085 (RIN64_23085) | 88142..88879 | + | 738 | WP_313830649 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| RIN64_RS23090 (RIN64_23090) | 88896..89141 | + | 246 | WP_102750541 | type-F conjugative transfer system pilin chaperone TraQ | - |
| RIN64_RS23095 (RIN64_23095) | 89131..89670 | + | 540 | WP_313830621 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| RIN64_RS23100 (RIN64_23100) | 89670..91043 | + | 1374 | WP_313830651 | conjugal transfer pilus assembly protein TraH | traH |
| RIN64_RS23105 (RIN64_23105) | 91043..93925 | + | 2883 | WP_313830623 | conjugal transfer mating-pair stabilization protein TraG | traG |
| RIN64_RS23110 (RIN64_23110) | 93937..94470 | + | 534 | WP_313830625 | hypothetical protein | - |
| RIN64_RS23115 (RIN64_23115) | 94835..97075 | + | 2241 | WP_313830627 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 16308 | GenBank | NZ_CP134405 |
| Plasmid name | 3.2.17|unnamed | Incompatibility group | IncFIB |
| Plasmid size | 105076 bp | Coordinate of oriT [Strand] | 74637..74749 [+] |
| Host baterium | Enterobacter cancerogenus strain 3.2.17 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA7 |