Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 115837 |
| Name | oriT_NZLR24|p4 |
| Organism | Rhizobium ruizarguesonis strain NZLR24 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP071682 (158964..158997 [-], 34 nt) |
| oriT length | 34 nt |
| IRs (inverted repeats) | 19..24, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 34 nt
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file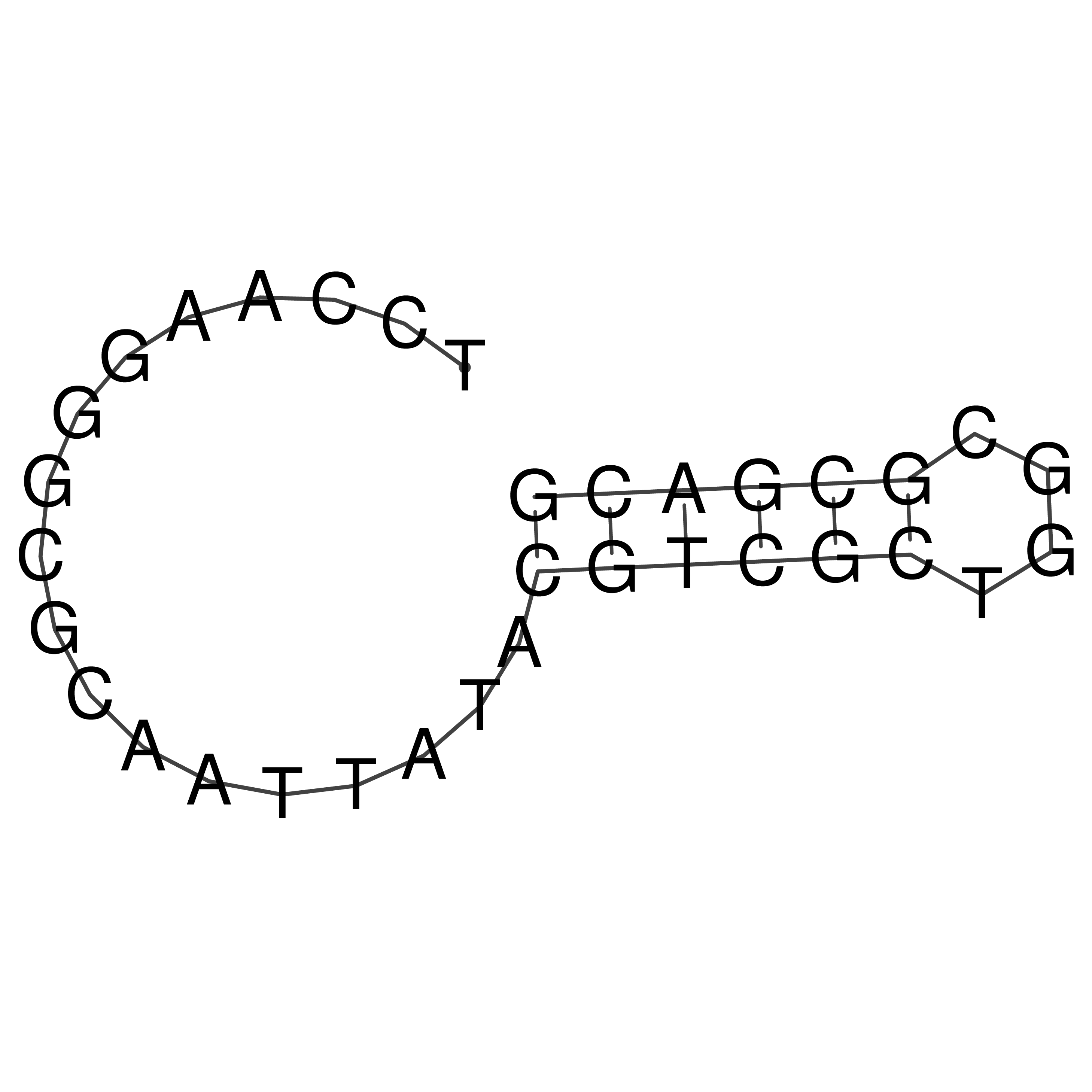
Relaxase
| ID | 10146 | GenBank | WP_207580365 |
| Name | traA_J3P73_RS35050_NZLR24|p4 |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 122803.82 Da Isoelectric Point: 9.7932
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVAGHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKPIRNDAGKIVYELWAGSTEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTREM
IRLEAEMANRAIWLSGRASLGVREAVLEATFARHSRLSDEQRTAIEHVAGATRIAAVIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNQLDDQTVFVLDEAGMVSSRQM
ALFVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRNASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRMLNNMARAKLVERGVVAHGFAF
KTEDGTRVFATGDQIVFLKNEGSLGVKNGMLAKVLEAVAGRIVAEIGDGEHRRQVTIEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETTLDYE
KASFYGQALRFAEARGLHLVNVARTIAHDRLQWAVRQSSRLADLGARLAAIGAALGLVRDSNKHSVPDTI
KETKPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDVSTRFQLVYAKPEAAFKAINVDTMLKDQSVAQS
TLARIAGEPESFGALKGKTGLLASRGDKQDREKALANVPALARNLERYLRERAEAELKHETEERAVRLKV
SVDIPALSPSAKRTLERVRDAIDRNDLPSGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTSGMTAGQKAEVRSAWNSMRTVQQLGSHERTTEAAKQAETMRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 11853 | GenBank | WP_207580341 |
| Name | t4cp2_J3P73_RS34465_NZLR24|p4 |
UniProt ID | _ |
| Length | 907 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 907 a.a. Molecular weight: 101280.51 Da Isoelectric Point: 8.2223
MASVSTHTETDRPKWTGLQRASLFTLPIALFGVVLLWLAFFTLIYDAYITSFGSSSYFDYVGQARLIDAV
RYTWSIIHVRNTVGLSLLAFVLALPFLSFMTCLWMLKGGRALSRKILYLLHIRSMFSLIAHTALIFAAAY
AGVIVYGVAFYLVATRAGEHAEIASYYGNHLAAMYQAPFGATDAAGAYQKPSRQTVLAALAGLAAAGAIV
GFPIVAAIQKRQRMIMGKARLATLKDAADFRLRDKHGIVLGLKQGLLLRNDGDQHVMVIGSPGQGKSRGF
VIPTMMSFQGSQVVLDMSGELFEETSGYLKDKGYEVFLLAPGSRYTDGYNPLDLISDEPNQRITDLQKLT
QMLLPERLRSDSSDFWEESARILLTAMLGFVLECPDTRKSLSELYRILNSMSDERKAIIQLLENYEPVLS
DQTRMQLTKFAGRHEKLGEGIAAEIVAKLNFLQNPLVEALTSITTIPIGTIRKRKLAIYIQADWNAMQIY
ERLISMFIQQMADKLVQMGPLRNGEHEVLMMLDEFGNGGRIDTVLTLAPLIRKNGVRFVFILQDGAQLER
LYQRSGQKILMGASTIKLFMNFQNQEDATAVSMAAGKTTEWVEQSSYSHRHGRRQRSISKVPVSVDLLPI
NTLMMMKPEEAILQVTGMPLMRIMKLDSGGERMFSKVRKFKPVKRPCMEPVEWTIADSRSAAAAPPATGA
HIHFDRSVVKDAVRHVYISSLDELRHRLDLDETREAGVTEKEAPEEEKKQPGGKRKGDPANGKRKRSMPK
TSGAARESDNDLNIDEEKLSRNVYSGSPAHRSVRSKQTNGPTDHIVPRPVAPVLQRQEVRDAFGEDLNAL
NDVIFQQAEENALEPVELNADVELLIESLSNRLTEDSSRAYLYQFANLARDPLGEDREQYETEAHAD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 11854 | GenBank | WP_164580028 |
| Name | virB11_J3P73_RS34470_NZLR24|p4 |
UniProt ID | _ |
| Length | 349 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 349 a.a. Molecular weight: 39031.31 Da Isoelectric Point: 7.1229
MNARPSISAEQHFLSEAIEPIRPFLDDPNVVEISCQRSNEIWLERIGQLSMVRQDVDGLDQDAIRHMASR
AASFTKQTVNAQHPLLSATLVNGERVQFVLNPTSATGHAFSIRKQFIRRFDLNDYEKAGAFRFTKVTGDD
YIDDVDIELSRLLRAGKVRAFLELAAINHRSMLISGGTSTGKTTFLNTMLTAIPNHERFILMQDTAELKP
RQENVVSLLISKGGQGEARVTMGDLLATALRLRPDRIFMGELRGDEAFDFLNAINTGHPGSMSTIHANSP
HHAFNRLSTLVLNSNVRMERDDIIRYIRSIIPIVVQLKKSDGAGGRGVSEIYFADEVDENGKRIHAHRD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 11855 | GenBank | WP_207580361 |
| Name | t4cp2_J3P73_RS34970_NZLR24|p4 |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 91350.99 Da Isoelectric Point: 5.5804
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERTELSRQINAVLSRLG
SGWMIQVDAIRIPTVDYPSEDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRTKSYADTVLFVFKNAVRELEQYFANTLSIRRMETRETVEGGGERIARYDELLQFARFCTTG
ESHPIRLPDIPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVAETEDAIAQASSQLVAYGYYTPVVVL
FDSDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYEHAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSDLKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTIDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLEGSPTLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAIPKREYYVATPEGRRLFDMALGP
VALSFVGASGKEDLKRIRGLKSEHGHEWPIHWLETRGIHDAASLLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 11856 | GenBank | WP_207580369 |
| Name | traG_J3P73_RS35090_NZLR24|p4 |
UniProt ID | _ |
| Length | 643 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 643 a.a. Molecular weight: 69246.37 Da Isoelectric Point: 9.2306
MTINRIALVVVPGTLMILVVIGMTGIEQWLSGFGKTEAARLAWGRTGIALPYVASAAIGILLLFSSAGSI
NIKQAGWGVVAGCSGTILIAAIRETMRLSAFMTVPADKTVWAFLDPATSIGASAALLCAGFALRVALIGN
AAFARAEPKRIQGKRALHGEAGWMKLTEAAKLFPDAGGIIIGERYRVDKDSVGSQAFRADSAETWGAGGK
SPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSGHRGGAGRDVFVLDP
RKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDGGGVRGVRDDFFRASALQLLTALIADVCLSGHTDGH
DQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYAA
LVSGKKFATNEIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNHDGQIKGRALFLLDEVARLGYM
RIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTTV
EIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRIRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVGT
NRFHMVGDTPKPA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 30965..41411
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| J3P73_RS34450 (J3P73_34450) | 26119..26412 | + | 294 | WP_164580024 | hypothetical protein | - |
| J3P73_RS34455 (J3P73_34455) | 26423..27811 | + | 1389 | WP_164580025 | hypothetical protein | - |
| J3P73_RS34460 (J3P73_34460) | 27815..28090 | - | 276 | WP_245002799 | hypothetical protein | - |
| J3P73_RS34465 (J3P73_34465) | 28273..30996 | - | 2724 | WP_207580341 | type IV secretory system conjugative DNA transfer family protein | - |
| J3P73_RS34470 (J3P73_34470) | 30965..32014 | - | 1050 | WP_164580028 | P-type DNA transfer ATPase VirB11 | virB11 |
| J3P73_RS34475 (J3P73_34475) | 32011..33018 | - | 1008 | WP_245002800 | type IV secretion system protein VirB10 | virB10 |
| J3P73_RS34480 (J3P73_34480) | 33359..34177 | - | 819 | WP_245002801 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| J3P73_RS34485 (J3P73_34485) | 34186..34920 | - | 735 | WP_164579990 | virB8 family protein | virB8 |
| J3P73_RS34490 (J3P73_34490) | 34949..36022 | - | 1074 | WP_164579989 | type IV secretion system protein | virB6 |
| J3P73_RS34495 (J3P73_34495) | 36231..37022 | - | 792 | WP_164579988 | conjugal transfer protein | - |
| J3P73_RS34500 (J3P73_34500) | 37046..38221 | - | 1176 | WP_164579987 | lytic transglycosylase domain-containing protein | - |
| J3P73_RS34505 (J3P73_34505) | 38199..40718 | - | 2520 | WP_164579986 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| J3P73_RS34510 (J3P73_34510) | 40722..41048 | - | 327 | WP_164579985 | VirB3 family type IV secretion system protein | virB3 |
| J3P73_RS34515 (J3P73_34515) | 41052..41411 | - | 360 | WP_164579984 | TrbC/VirB2 family protein | virB2 |
| J3P73_RS34520 (J3P73_34520) | 41411..42484 | - | 1074 | WP_164579983 | lytic transglycosylase domain-containing protein | - |
| J3P73_RS34525 (J3P73_34525) | 42650..43153 | - | 504 | WP_164579982 | hypothetical protein | - |
| J3P73_RS37855 | 43165..43659 | - | 495 | WP_245002802 | hypothetical protein | - |
| J3P73_RS37860 | 44333..44515 | + | 183 | WP_245002803 | hypothetical protein | - |
| J3P73_RS37865 | 44626..44724 | + | 99 | Protein_50 | LysR family transcriptional regulator | - |
| J3P73_RS34535 (J3P73_34535) | 45785..46267 | - | 483 | WP_164579981 | hypothetical protein | - |
Region 2: 140028..153828
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| J3P73_RS34935 (J3P73_34935) | 135355..136569 | - | 1215 | WP_207580407 | plasmid replication protein RepC | - |
| J3P73_RS34940 (J3P73_34940) | 136759..137796 | - | 1038 | WP_164021417 | plasmid partitioning protein RepB | - |
| J3P73_RS34945 (J3P73_34945) | 137793..139016 | - | 1224 | WP_032991837 | plasmid partitioning protein RepA | - |
| J3P73_RS34950 (J3P73_34950) | 139393..140031 | + | 639 | WP_131624136 | acyl-homoserine-lactone synthase | - |
| J3P73_RS34955 (J3P73_34955) | 140028..141005 | + | 978 | WP_131624138 | P-type conjugative transfer ATPase TrbB | virB11 |
| J3P73_RS34960 (J3P73_34960) | 140995..141378 | + | 384 | WP_164021416 | TrbC/VirB2 family protein | virB2 |
| J3P73_RS34965 (J3P73_34965) | 141371..141670 | + | 300 | WP_027668110 | conjugal transfer protein TrbD | virB3 |
| J3P73_RS34970 (J3P73_34970) | 141681..144131 | + | 2451 | WP_207580361 | conjugal transfer protein TrbE | virb4 |
| J3P73_RS34975 (J3P73_34975) | 144109..144906 | + | 798 | WP_131624147 | P-type conjugative transfer protein TrbJ | virB5 |
| J3P73_RS34980 (J3P73_34980) | 144903..145103 | + | 201 | WP_207580408 | entry exclusion protein TrbK | - |
| J3P73_RS34985 (J3P73_34985) | 145097..146278 | + | 1182 | WP_207580362 | P-type conjugative transfer protein TrbL | virB6 |
| J3P73_RS34990 (J3P73_34990) | 146299..146961 | + | 663 | WP_131624151 | conjugal transfer protein TrbF | virB8 |
| J3P73_RS34995 (J3P73_34995) | 146979..147791 | + | 813 | WP_131624153 | P-type conjugative transfer protein TrbG | virB9 |
| J3P73_RS35000 (J3P73_35000) | 147795..148235 | + | 441 | WP_131624155 | conjugal transfer protein TrbH | - |
| J3P73_RS35005 (J3P73_35005) | 148247..149545 | + | 1299 | WP_207580363 | IncP-type conjugal transfer protein TrbI | virB10 |
| J3P73_RS35010 (J3P73_35010) | 149856..150560 | + | 705 | WP_077988209 | autoinducer binding domain-containing protein | - |
| J3P73_RS35015 (J3P73_35015) | 150564..150854 | - | 291 | WP_077988208 | transcriptional repressor TraM | - |
| J3P73_RS35020 (J3P73_35020) | 151114..151362 | + | 249 | WP_027668121 | helix-turn-helix domain-containing protein | - |
| J3P73_RS35025 (J3P73_35025) | 151605..152222 | + | 618 | WP_077988207 | type IV toxin-antitoxin system AbiEi family antitoxin domain-containing protein | - |
| J3P73_RS35030 (J3P73_35030) | 152215..153080 | + | 866 | Protein_149 | nucleotidyl transferase AbiEii/AbiGii toxin family protein | - |
| J3P73_RS35035 (J3P73_35035) | 153211..153828 | - | 618 | WP_164021400 | TraH family protein | virB1 |
| J3P73_RS35040 (J3P73_35040) | 153845..155008 | - | 1164 | WP_168334974 | conjugal transfer protein TraB | - |
| J3P73_RS35045 (J3P73_35045) | 154998..155561 | - | 564 | WP_207580364 | conjugative transfer signal peptidase TraF | - |
Host bacterium
| ID | 16270 | GenBank | NZ_CP071682 |
| Plasmid name | NZLR24|p4 | Incompatibility group | - |
| Plasmid size | 643193 bp | Coordinate of oriT [Strand] | 158964..158997 [-] |
| Host baterium | Rhizobium ruizarguesonis strain NZLR24 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | hupE |
| Degradation gene | - |
| Symbiosis gene | nodM, nodE, nodF, nodD, nodA, nodC, nodI, nodJ, nifB, fixX, fixC, fixB, fixA, nifH, nifD, nifE, nifN, fixQ, fixO, fixN |
| Anti-CRISPR | - |