Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 115814 |
| Name | oriT_54A:C|unnamed2 |
| Organism | Enterobacter ludwigii strain 54A:C |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP134656 (73065..73151 [-], 87 nt) |
| oriT length | 87 nt |
| IRs (inverted repeats) | 28..35, 38..45 (GCAAAATT..AATTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 87 nt
>oriT_54A:C|unnamed2
CTCACCACCAAAAGCACCACACCCCACGCAAAATTAAAATTTTGCTGATTGGCTCTTTAAATCATGCAGTTAGTTAAAAAGTAATGT
CTCACCACCAAAAGCACCACACCCCACGCAAAATTAAAATTTTGCTGATTGGCTCTTTAAATCATGCAGTTAGTTAAAAAGTAATGT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file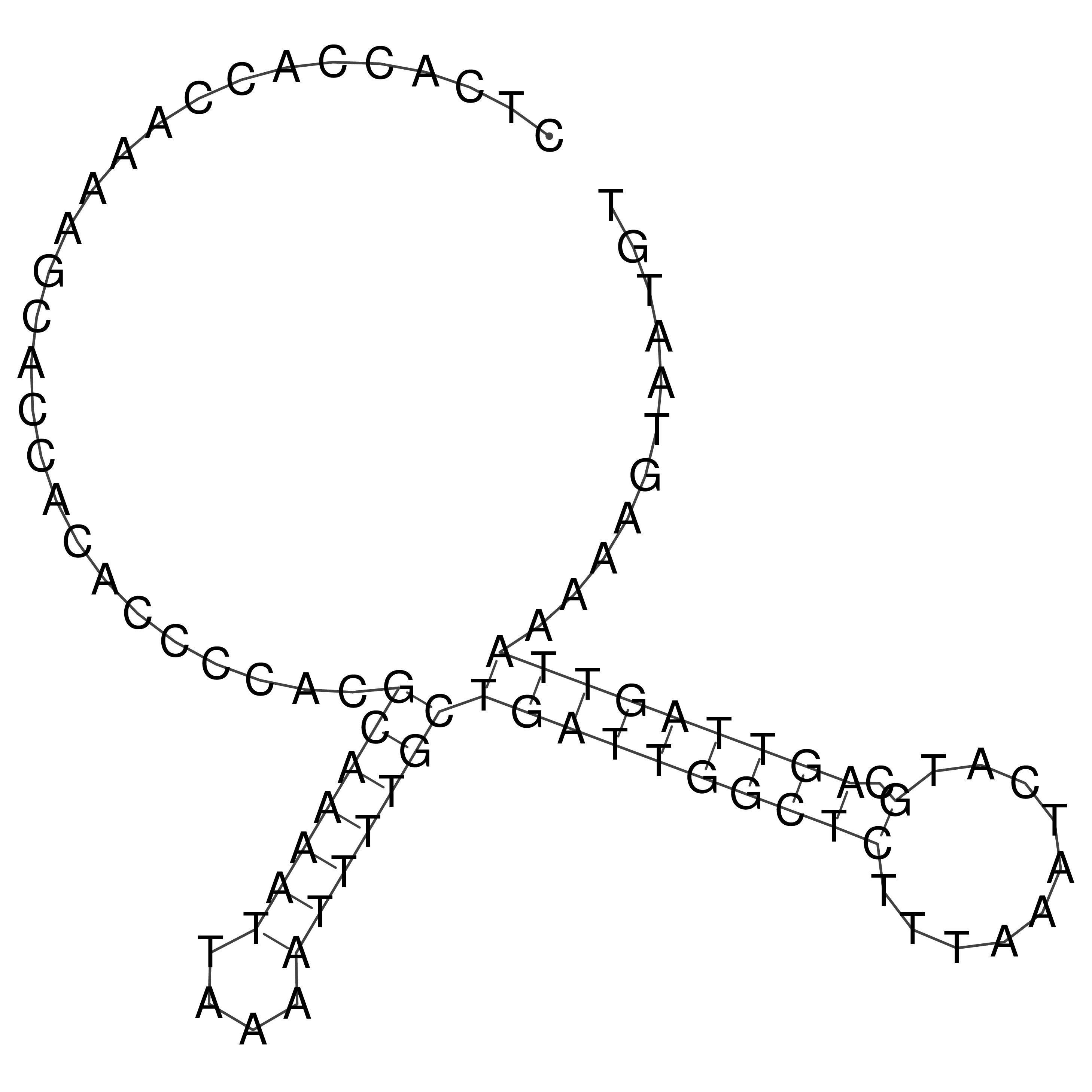
Relaxase
| ID | 10133 | GenBank | WP_311382035 |
| Name | traI_RIK68_RS24890_54A:C|unnamed2 |
UniProt ID | _ |
| Length | 1746 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1746 a.a. Molecular weight: 189200.57 Da Isoelectric Point: 6.2072
>WP_311382035.1 conjugative transfer relaxase/helicase TraI [Enterobacter ludwigii]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGKGAEALGLSGGVDQKVFTKVLEGRLPDGSDLSRTQ
DGANKHRPGYDFTFSAPKSVSVMAMLGGDKRLIEAHNRAVETAIKQIEAMASTRVMTEGRSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANATQHGDEWRTLSSDKVGKTGFIENIYANQIAFGRLYRAALKDDVAA
MGYDTETVGKHGMWELKGVPTEPYSSRSRTISEAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMQT
LKATGFDIKAYREAADLRVEQGNIPVTAPEAIDINSSVGQSIAMLSDRRARFTYSELLATTLGQLPARPG
MVEMARDGIDAAIKNEQLIPLDKEKGLFTSNIHVLDELSVSAMTRDLQRAGKADIFPEKGVPRSRSYSDA
VSVLAQDRPPVAIISGQGGAAGQRERVAELTMMAREQGRDVHIIAADRRSRSALMQDERLSGEHITDRRG
LTEGMTFTPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGLSTLAWQG
GEKTQVSVISEPDRRQRYDRLAADFARSVRAGEHSVAQVTGPREQAVLAGVVREALKAEKVLGEREVSIT
TLEPVWLDSRHQQVRDHYREGMVMERWNAEERSRERFVIDRVTGRNNSLTLRNAQGETRVTRLSELNSSW
SLYRTGMLHVAEGDRLAVLGQAQGTRLKGGDTVTVKALGEAGLVVSLPGRKADVVLPAGDSPFTAAKVGQ
GWVESPGRSVSDSATVFASLSQRELDNTTLNKLALSGRAVRLYSAQTAQKTTEKLSRQSAWSVVSEQIKD
AAGHDRLDDALAHQKAGLHTPEQQAIHLAIPVLEGNGLAFTKPQLMAAAKEFEQDSLSLQAIEKEADRQV
RSGALLSVPVSPGNGLQLLVSRQSYNAEKSIIRHVLEGKEAVTPLMDKVPDAQLAGLTDGQRNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEREQPRVIGVAPTHRAVSEMRDAGVPETQTLAAFIHDTQ
QQLRGGERPDFSNVLFLVDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DVAVMQEIVRQTPELKPAVYSLINRDIGSALTTIERVAPAQVPRRADAWQPDSSVMEFSREREEAIAKAV
AAGDLMPGGQPATLLEAIVKDYTGRTPEAQAQTIVITALNADRRQVNAMIHDARQDAGEVGEKEVTLPVL
TPANIRDGELRRMTTWEASRDSLVLLDSTYYSIEALDGEAHLVTLKDAQGNTRSLSPAQAATEGVTLYRQ
DTITVSEGDRMRFSKSDNERGFVANSVWSVSEIKGDSVTLSDGKQTRTLTPSAEQAEQHIDLAYAVTVEG
SQGASEPFSISLQGTDGGRKRMVSFESAYVALSRMKQHAQVYTDNRDKWVAAMEKSQAKSTAHDILEPRG
DRAVANAARLTATAKALGEVPAGLAALSQIGLQPEGSMAKFISPGRKYPQPHVALPAFDRNGRQAGVWLS
ALTSGDGQLKGLGGEGRVMGSGDAAFAGLQASRNGESLLARDMEEGVRLARENPQSGVVVRLVGEARPWN
PGAITGGRVWADSLPDSTGTQPGETVPPEVLAQQAREAEVQRELDKRAEEAVREMARSGDGPAGGADVAV
RDVVRDMKRIKETPVSPLTLPDAPEERRRDEAVSQVVRENVQRDRLQQMERETVRDLEREKTLGGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGKGAEALGLSGGVDQKVFTKVLEGRLPDGSDLSRTQ
DGANKHRPGYDFTFSAPKSVSVMAMLGGDKRLIEAHNRAVETAIKQIEAMASTRVMTEGRSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANATQHGDEWRTLSSDKVGKTGFIENIYANQIAFGRLYRAALKDDVAA
MGYDTETVGKHGMWELKGVPTEPYSSRSRTISEAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMQT
LKATGFDIKAYREAADLRVEQGNIPVTAPEAIDINSSVGQSIAMLSDRRARFTYSELLATTLGQLPARPG
MVEMARDGIDAAIKNEQLIPLDKEKGLFTSNIHVLDELSVSAMTRDLQRAGKADIFPEKGVPRSRSYSDA
VSVLAQDRPPVAIISGQGGAAGQRERVAELTMMAREQGRDVHIIAADRRSRSALMQDERLSGEHITDRRG
LTEGMTFTPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGLSTLAWQG
GEKTQVSVISEPDRRQRYDRLAADFARSVRAGEHSVAQVTGPREQAVLAGVVREALKAEKVLGEREVSIT
TLEPVWLDSRHQQVRDHYREGMVMERWNAEERSRERFVIDRVTGRNNSLTLRNAQGETRVTRLSELNSSW
SLYRTGMLHVAEGDRLAVLGQAQGTRLKGGDTVTVKALGEAGLVVSLPGRKADVVLPAGDSPFTAAKVGQ
GWVESPGRSVSDSATVFASLSQRELDNTTLNKLALSGRAVRLYSAQTAQKTTEKLSRQSAWSVVSEQIKD
AAGHDRLDDALAHQKAGLHTPEQQAIHLAIPVLEGNGLAFTKPQLMAAAKEFEQDSLSLQAIEKEADRQV
RSGALLSVPVSPGNGLQLLVSRQSYNAEKSIIRHVLEGKEAVTPLMDKVPDAQLAGLTDGQRNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEREQPRVIGVAPTHRAVSEMRDAGVPETQTLAAFIHDTQ
QQLRGGERPDFSNVLFLVDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DVAVMQEIVRQTPELKPAVYSLINRDIGSALTTIERVAPAQVPRRADAWQPDSSVMEFSREREEAIAKAV
AAGDLMPGGQPATLLEAIVKDYTGRTPEAQAQTIVITALNADRRQVNAMIHDARQDAGEVGEKEVTLPVL
TPANIRDGELRRMTTWEASRDSLVLLDSTYYSIEALDGEAHLVTLKDAQGNTRSLSPAQAATEGVTLYRQ
DTITVSEGDRMRFSKSDNERGFVANSVWSVSEIKGDSVTLSDGKQTRTLTPSAEQAEQHIDLAYAVTVEG
SQGASEPFSISLQGTDGGRKRMVSFESAYVALSRMKQHAQVYTDNRDKWVAAMEKSQAKSTAHDILEPRG
DRAVANAARLTATAKALGEVPAGLAALSQIGLQPEGSMAKFISPGRKYPQPHVALPAFDRNGRQAGVWLS
ALTSGDGQLKGLGGEGRVMGSGDAAFAGLQASRNGESLLARDMEEGVRLARENPQSGVVVRLVGEARPWN
PGAITGGRVWADSLPDSTGTQPGETVPPEVLAQQAREAEVQRELDKRAEEAVREMARSGDGPAGGADVAV
RDVVRDMKRIKETPVSPLTLPDAPEERRRDEAVSQVVRENVQRDRLQQMERETVRDLEREKTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 11838 | GenBank | WP_311382034 |
| Name | traD_RIK68_RS24895_54A:C|unnamed2 |
UniProt ID | _ |
| Length | 734 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 734 a.a. Molecular weight: 82090.63 Da Isoelectric Point: 5.1609
>WP_311382034.1 type IV conjugative transfer system coupling protein TraD [Enterobacter ludwigii]
MSFNAKDMTQGGQIASMRIRMFSQIANIIVYCLVILFGVIVAITLSVKLTWQTFVNGMVYYWCQTLAPMR
DIFKSQPVYTIRYYGHELKYNAQQILQDKYIQWCADQVWTAFILAAIIAAVVCVITFFVTTWILGHQGKQ
QSEDEITGGRQLTDDPKAVAKMLRRDGLASDIKIGELPLIKDSEIQNFCLHGTVGAGKSEVIRRLMNYAR
QRGDMVIVYDRSCEFVKSYYDPATDKILNPLDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLKNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIENNGESFTIRDWMRGVREDGKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDVQTLPDLSCYVTLPGPYPAVKMAL
KYQQRPKVAPEFIPRTIDGKAEERLNAALAAREHESRSIAMLFEPDADVIAPEGGAENAGQPQQPQQPQQ
PQQPQQPQQLAPAPRAADTASASASAAGAGGAGGVEPELKTKAEEAEQLPPGISESGEVVDMAAWEAWQA
EQDELSPLERQRREEVNINVHRAPEKDLEPGGYI
MSFNAKDMTQGGQIASMRIRMFSQIANIIVYCLVILFGVIVAITLSVKLTWQTFVNGMVYYWCQTLAPMR
DIFKSQPVYTIRYYGHELKYNAQQILQDKYIQWCADQVWTAFILAAIIAAVVCVITFFVTTWILGHQGKQ
QSEDEITGGRQLTDDPKAVAKMLRRDGLASDIKIGELPLIKDSEIQNFCLHGTVGAGKSEVIRRLMNYAR
QRGDMVIVYDRSCEFVKSYYDPATDKILNPLDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLKNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIENNGESFTIRDWMRGVREDGKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDVQTLPDLSCYVTLPGPYPAVKMAL
KYQQRPKVAPEFIPRTIDGKAEERLNAALAAREHESRSIAMLFEPDADVIAPEGGAENAGQPQQPQQPQQ
PQQPQQPQQLAPAPRAADTASASASAAGAGGAGGVEPELKTKAEEAEQLPPGISESGEVVDMAAWEAWQA
EQDELSPLERQRREEVNINVHRAPEKDLEPGGYI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 11839 | GenBank | WP_063925869 |
| Name | traC_RIK68_RS24970_54A:C|unnamed2 |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 98769.44 Da Isoelectric Point: 5.7676
>WP_063925869.1 type IV secretion system protein TraC [Enterobacter ludwigii]
MSNPLDTVTRTVNALVTALKMPDESAKANDVLGQMSFPQFSRVLPYRDYDAETGLFMNEKTMGFMLEAQP
INGANETIVSSLENLMRTKLPREVPVAVHLVSSHLVGQDIEYGLRELSWTGEKADTFNAITRAYYLRAAE
EGFPLPDHLNLPLTLRNYRVYISCCVPRKRGSKASVMEMENQVKIIRASLQGAQIATRSVDADAFIRIAG
EMINHNPDALYPTERELDPLKDLNYQCVDDSFDLKVKADYMTLGLRGNGRNSTARIMNFQLTKNPELAFL
WNMADNYSNLLNPELSVSCPFILTLVLSVEDQVKAQNEANLKFMDLEKKSKTSYAKFFPGVAQEAKEWGD
LRQRLSSGQSSLVSYFLNVTLFCRDDQETALKVEQDVLNSFRKNGFELISPRFNHMRNFLVNLPFMAGGG
LFKQLRESGVVQRAESFNVANLMPVVADSPLATSGLLAPTYRNQLAFIDLFSRGMGNTNFNMAVCGTSGA
GKTGLIQPLIRSVIDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLKFNPFANVTDDTIDEMAERLRDQL
SVMASPNGNLDEVHEGLLLKAVKATWLTKKNQARIDDVVDYLMMARDSEEYSGSPTIRSRLDEMVVLLTQ
YTRDGVYGSYFNSDEPSLKDEARMVVLELGGLEDRPSLLVAVMFSLILYIENRMYRTPRSLKKLNVIDEG
WRLLDFKNNKVGAFIEKGYRTARRHTGSYITITQNIVDFDSETASSAARAAWGNSSYKIILKQSAKEFAK
YNQLYPDQYSQLQRDMIKGFGSAKDNWFSSFMLSVEHLTSWHRLFVDPLSRAMYSSDGPDFEFLQQRRRE
GLSIHEAVWELAWKKSGPEMAALERWLHEHEGKTIS
MSNPLDTVTRTVNALVTALKMPDESAKANDVLGQMSFPQFSRVLPYRDYDAETGLFMNEKTMGFMLEAQP
INGANETIVSSLENLMRTKLPREVPVAVHLVSSHLVGQDIEYGLRELSWTGEKADTFNAITRAYYLRAAE
EGFPLPDHLNLPLTLRNYRVYISCCVPRKRGSKASVMEMENQVKIIRASLQGAQIATRSVDADAFIRIAG
EMINHNPDALYPTERELDPLKDLNYQCVDDSFDLKVKADYMTLGLRGNGRNSTARIMNFQLTKNPELAFL
WNMADNYSNLLNPELSVSCPFILTLVLSVEDQVKAQNEANLKFMDLEKKSKTSYAKFFPGVAQEAKEWGD
LRQRLSSGQSSLVSYFLNVTLFCRDDQETALKVEQDVLNSFRKNGFELISPRFNHMRNFLVNLPFMAGGG
LFKQLRESGVVQRAESFNVANLMPVVADSPLATSGLLAPTYRNQLAFIDLFSRGMGNTNFNMAVCGTSGA
GKTGLIQPLIRSVIDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLKFNPFANVTDDTIDEMAERLRDQL
SVMASPNGNLDEVHEGLLLKAVKATWLTKKNQARIDDVVDYLMMARDSEEYSGSPTIRSRLDEMVVLLTQ
YTRDGVYGSYFNSDEPSLKDEARMVVLELGGLEDRPSLLVAVMFSLILYIENRMYRTPRSLKKLNVIDEG
WRLLDFKNNKVGAFIEKGYRTARRHTGSYITITQNIVDFDSETASSAARAAWGNSSYKIILKQSAKEFAK
YNQLYPDQYSQLQRDMIKGFGSAKDNWFSSFMLSVEHLTSWHRLFVDPLSRAMYSSDGPDFEFLQQRRRE
GLSIHEAVWELAWKKSGPEMAALERWLHEHEGKTIS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 48636..73699
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| RIK68_RS24895 (RIK68_24895) | 48636..50840 | - | 2205 | WP_311382034 | type IV conjugative transfer system coupling protein TraD | virb4 |
| RIK68_RS24900 (RIK68_24900) | 51015..51746 | - | 732 | WP_063925860 | conjugal transfer complement resistance protein TraT | - |
| RIK68_RS24905 (RIK68_24905) | 51840..52151 | + | 312 | WP_063925861 | heavy metal-binding domain-containing protein | - |
| RIK68_RS24910 (RIK68_24910) | 52153..55011 | - | 2859 | WP_063925862 | conjugal transfer mating-pair stabilization protein TraG | traG |
| RIK68_RS24915 (RIK68_24915) | 55011..56381 | - | 1371 | WP_059514056 | conjugal transfer pilus assembly protein TraH | traH |
| RIK68_RS24920 (RIK68_24920) | 56368..56796 | - | 429 | WP_063925863 | hypothetical protein | - |
| RIK68_RS24925 (RIK68_24925) | 56789..57343 | - | 555 | WP_063925864 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| RIK68_RS24930 (RIK68_24930) | 57333..57581 | - | 249 | WP_008324126 | type-F conjugative transfer system pilin chaperone TraQ | - |
| RIK68_RS24935 (RIK68_24935) | 57598..58347 | - | 750 | WP_063925865 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| RIK68_RS24940 (RIK68_24940) | 58378..58566 | - | 189 | WP_016674191 | hypothetical protein | - |
| RIK68_RS24945 (RIK68_24945) | 58598..60478 | - | 1881 | WP_063925866 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| RIK68_RS24950 (RIK68_24950) | 60475..61104 | - | 630 | WP_063925867 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| RIK68_RS24955 (RIK68_24955) | 61121..62107 | - | 987 | WP_063925890 | conjugal transfer pilus assembly protein TraU | traU |
| RIK68_RS24960 (RIK68_24960) | 62110..62757 | - | 648 | WP_008324132 | type-F conjugative transfer system protein TraW | traW |
| RIK68_RS24965 (RIK68_24965) | 62757..63104 | - | 348 | WP_063925868 | type-F conjugative transfer system protein TrbI | - |
| RIK68_RS24970 (RIK68_24970) | 63101..65731 | - | 2631 | WP_063925869 | type IV secretion system protein TraC | virb4 |
| RIK68_RS24975 (RIK68_24975) | 65724..66290 | - | 567 | WP_063925870 | conjugal transfer protein TraP | - |
| RIK68_RS24980 (RIK68_24980) | 66280..66576 | - | 297 | WP_063925891 | hypothetical protein | - |
| RIK68_RS24985 (RIK68_24985) | 66749..66973 | - | 225 | WP_016674195 | TraR/DksA C4-type zinc finger protein | - |
| RIK68_RS24990 (RIK68_24990) | 67093..67632 | - | 540 | WP_063925871 | type IV conjugative transfer system lipoprotein TraV | traV |
| RIK68_RS24995 (RIK68_24995) | 67654..69048 | - | 1395 | WP_063925872 | F-type conjugal transfer pilus assembly protein TraB | traB |
| RIK68_RS25000 (RIK68_25000) | 69035..69778 | - | 744 | WP_063925873 | type-F conjugative transfer system secretin TraK | traK |
| RIK68_RS25005 (RIK68_25005) | 69765..70331 | - | 567 | WP_025759824 | type IV conjugative transfer system protein TraE | traE |
| RIK68_RS25010 (RIK68_25010) | 70346..70651 | - | 306 | WP_008324160 | type IV conjugative transfer system protein TraL | traL |
| RIK68_RS25015 (RIK68_25015) | 70802..71152 | - | 351 | WP_058610053 | type IV conjugative transfer system pilin TraA | - |
| RIK68_RS25020 (RIK68_25020) | 71210..71434 | - | 225 | WP_008324165 | TraY domain-containing protein | - |
| RIK68_RS25025 (RIK68_25025) | 71521..72210 | - | 690 | WP_063925874 | hypothetical protein | - |
| RIK68_RS25030 (RIK68_25030) | 72399..72791 | - | 393 | WP_063925875 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| RIK68_RS25035 (RIK68_25035) | 73223..73699 | + | 477 | WP_063925876 | transglycosylase SLT domain-containing protein | virB1 |
| RIK68_RS25040 (RIK68_25040) | 73746..74276 | - | 531 | WP_311382033 | antirestriction protein | - |
| RIK68_RS25045 (RIK68_25045) | 74903..75733 | - | 831 | WP_063925878 | N-6 DNA methylase | - |
| RIK68_RS25050 (RIK68_25050) | 75776..76039 | - | 264 | WP_063925879 | hypothetical protein | - |
| RIK68_RS25055 (RIK68_25055) | 76125..76469 | - | 345 | WP_047359689 | hypothetical protein | - |
| RIK68_RS25060 (RIK68_25060) | 76516..76806 | - | 291 | WP_240474975 | hypothetical protein | - |
| RIK68_RS25065 (RIK68_25065) | 76857..77231 | - | 375 | WP_032637252 | hypothetical protein | - |
| RIK68_RS25070 (RIK68_25070) | 77877..78251 | - | 375 | WP_063925880 | hypothetical protein | - |
| RIK68_RS25075 (RIK68_25075) | 78313..78636 | - | 324 | WP_063925881 | hypothetical protein | - |
Host bacterium
| ID | 16247 | GenBank | NZ_CP134656 |
| Plasmid name | 54A:C|unnamed2 | Incompatibility group | IncFII |
| Plasmid size | 163991 bp | Coordinate of oriT [Strand] | 73065..73151 [-] |
| Host baterium | Enterobacter ludwigii strain 54A:C |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA7 |