Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 115788 |
| Name | oriT_pWSM1592_2 |
| Organism | Rhizobium sullae strain WSM1592 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP104145 (450266..450299 [+], 34 nt) |
| oriT length | 34 nt |
| IRs (inverted repeats) | 19..24, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 34 nt
>oriT_pWSM1592_2
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACG
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file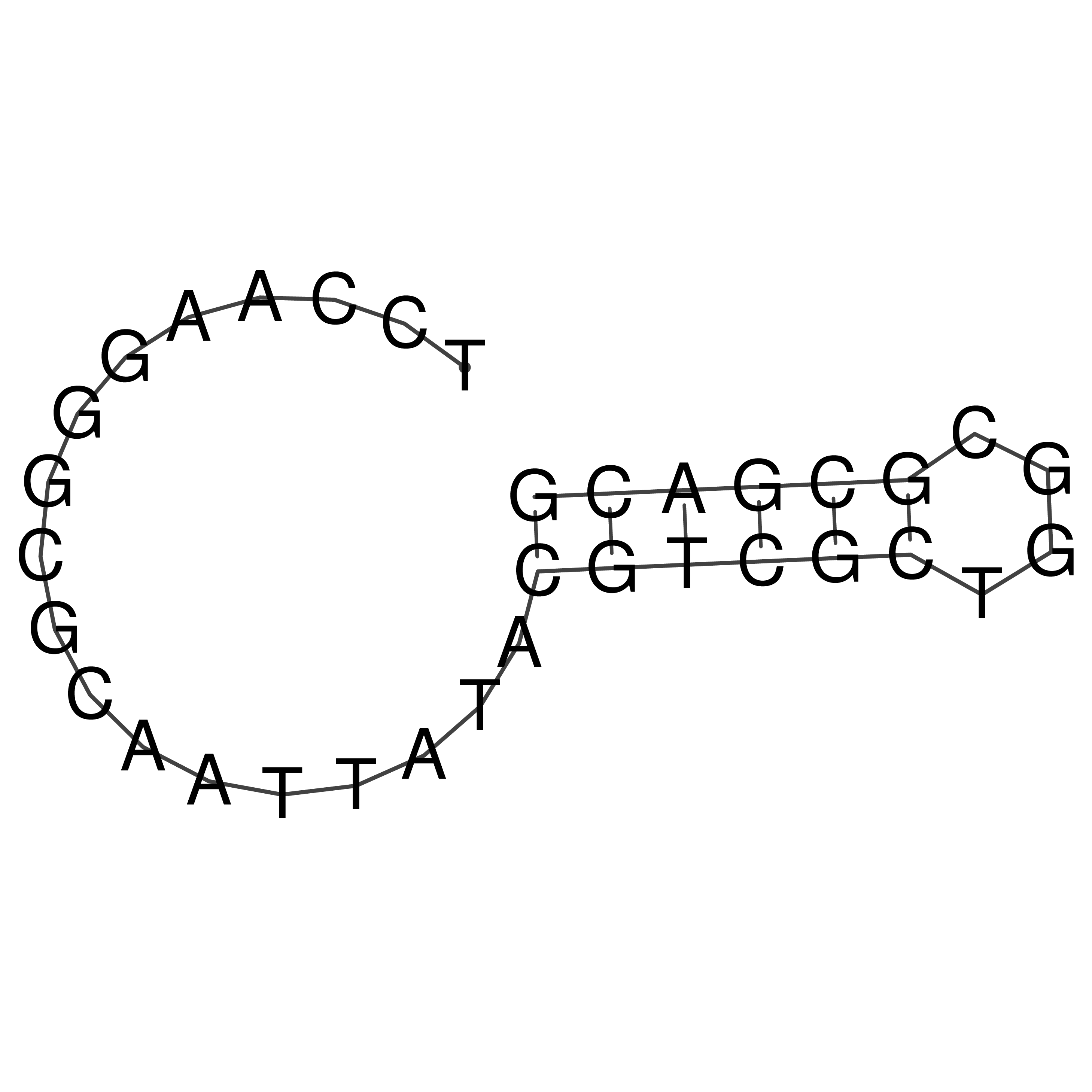
Relaxase
| ID | 10112 | GenBank | WP_027513360 |
| Name | traA_N2599_RS36320_pWSM1592_2 |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 122779.64 Da Isoelectric Point: 9.5327
>WP_027513360.1 Ti-type conjugative transfer relaxase TraA [Rhizobium sullae]
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRNMIADH
SVAGASEAFWNKAEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVEQHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLTENGFGSKKVAVLGPDGKPIRNDAGKIVYQLWAGCTEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQERRRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDAALFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTREM
IRLEAEMANRAIWLSGRASHDLREAVLQATLARHARLSDEQRTAIEHVAGGERIAAVVGRAGAGKTTMMR
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIQSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMASSRQM
ALFVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELDTIYRQRQQWMRDASLDLARGNVGKA
VDAYRAHGHVRGLDLKAQAVESLIADWDREYDPSKTSLILAHLRRDVRMLNEMARAKLVERGVIADGFAF
KTEDGTRMFATGDQIVFLKNEGSLGVKNGMLAKVLEAAPGRIVAEIGEGEHRRQVTIEQRFYNNLDHGYA
TSIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETTLDYE
KASFYGQALRFAEARGLHLVNVARTIAHDRLQWAVRQKQKLADLGARLAAIGAALGLVRGSNKHSIPDTI
KEAKPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDVSTRFQLVYAKPEAAFKAINVDAMVKDQSVAQS
TLARIAGEPESFGALNGKTGLLASRADKQDREKAVANVAALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVHDAIDRNDLPAGVEYALADKMVKAELEGFAKAVSERFGERTFLPLSAKDTDG
KTFETVTAGMTAGQKAEVQSAWNSMRTVQQLAAYERTTEALKHAETLRQTKSQGLSLK
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRNMIADH
SVAGASEAFWNKAEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVEQHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLTENGFGSKKVAVLGPDGKPIRNDAGKIVYQLWAGCTEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQERRRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDAALFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTREM
IRLEAEMANRAIWLSGRASHDLREAVLQATLARHARLSDEQRTAIEHVAGGERIAAVVGRAGAGKTTMMR
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIQSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMASSRQM
ALFVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELDTIYRQRQQWMRDASLDLARGNVGKA
VDAYRAHGHVRGLDLKAQAVESLIADWDREYDPSKTSLILAHLRRDVRMLNEMARAKLVERGVIADGFAF
KTEDGTRMFATGDQIVFLKNEGSLGVKNGMLAKVLEAAPGRIVAEIGEGEHRRQVTIEQRFYNNLDHGYA
TSIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETTLDYE
KASFYGQALRFAEARGLHLVNVARTIAHDRLQWAVRQKQKLADLGARLAAIGAALGLVRGSNKHSIPDTI
KEAKPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDVSTRFQLVYAKPEAAFKAINVDAMVKDQSVAQS
TLARIAGEPESFGALNGKTGLLASRADKQDREKAVANVAALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVHDAIDRNDLPAGVEYALADKMVKAELEGFAKAVSERFGERTFLPLSAKDTDG
KTFETVTAGMTAGQKAEVQSAWNSMRTVQQLAAYERTTEALKHAETLRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 11815 | GenBank | WP_037144402 |
| Name | virD4_N2599_RS35200_pWSM1592_2 |
UniProt ID | _ |
| Length | 571 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 571 a.a. Molecular weight: 63052.21 Da Isoelectric Point: 8.2537
>WP_037144402.1 type IV secretion system ATPase VirD4 [Rhizobium sullae]
MSSKTTFSNKTTPSSIALSILCSLAAGFCAASLYATFRHGISGEALMAFDILAFWYETPFYLGYTTLAFY
YGLAVVVSTSGIVLLVQQMLSEGSREHHGTARWAGVGEMRRTGYLQRYSRISGPIFGKTSGPFWSDYYLT
NGEQPHSLIVAPTRAGKGVGIVIPTLLTFKGSVIALDVKGELFELTSRARKADGDSVLKFAPLDPERRTS
CYNPLLDIIALPPERQFTEARRLAANLITTKGQGAEGFINGARDLFVAGILACIERGTPTIGAVYDLFAQ
PGEKYRLFARLAEETQNKEAQRIFDDMAGNDTKILTSYTSVLGDGGLNLWADPLVKAATSRSDFSIYDLR
RRRTCIYLCVSPNDLEVVAPLMRLLFQQVVSILQRSLPGKDEKHEVLFLLDEFKHLGKLEAVETAITTIA
GYKGRFMFIIQSLSALSGTYDEAGKQNFLSNTGVQVFMATADDETPNYISKAIGEYTFQARSTSHTQGRM
FDRNIQISDQGSPLLRPEQVRLLDDKCQIVLIKGQPPLQLRKVRYYSDRALKRIFESQTGGLPEPARLII
TEEGLGANCQS
MSSKTTFSNKTTPSSIALSILCSLAAGFCAASLYATFRHGISGEALMAFDILAFWYETPFYLGYTTLAFY
YGLAVVVSTSGIVLLVQQMLSEGSREHHGTARWAGVGEMRRTGYLQRYSRISGPIFGKTSGPFWSDYYLT
NGEQPHSLIVAPTRAGKGVGIVIPTLLTFKGSVIALDVKGELFELTSRARKADGDSVLKFAPLDPERRTS
CYNPLLDIIALPPERQFTEARRLAANLITTKGQGAEGFINGARDLFVAGILACIERGTPTIGAVYDLFAQ
PGEKYRLFARLAEETQNKEAQRIFDDMAGNDTKILTSYTSVLGDGGLNLWADPLVKAATSRSDFSIYDLR
RRRTCIYLCVSPNDLEVVAPLMRLLFQQVVSILQRSLPGKDEKHEVLFLLDEFKHLGKLEAVETAITTIA
GYKGRFMFIIQSLSALSGTYDEAGKQNFLSNTGVQVFMATADDETPNYISKAIGEYTFQARSTSHTQGRM
FDRNIQISDQGSPLLRPEQVRLLDDKCQIVLIKGQPPLQLRKVRYYSDRALKRIFESQTGGLPEPARLII
TEEGLGANCQS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 11816 | GenBank | WP_027513377 |
| Name | t4cp2_N2599_RS36405_pWSM1592_2 |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 91854.61 Da Isoelectric Point: 5.7886
>WP_027513377.1 conjugal transfer protein TrbE [Rhizobium sullae]
MVALKRFRATGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAILSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKTTALSK
YIYSDEESRKKSYADTVLFIFKNAVREIEQYFANTLSVRRMETRETVERGGERIARYDELLQFVRFCITG
ESHPIRLPDVPMYLDWIATAELEHGLTPKIESRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQLFQTQSRSFDQDAMTMVTETEDAIAQASSQLVAYGYYTPVVVL
FDHDREALQEKAEAIRRLIQAEGFGARIETLNATDAFLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNTVAPCPFYPPNSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYESAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAGEGRALAFCPLSDLKSDADRAWATEWIEMLVGLQRVTITPDHRN
AISRQVGLMASAAGRSLSDFVSGVQMREIKDALHHYTVDGPMGLLLDAEEDGLSLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRSKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNVAAREPGTREFYERIGFNERQIEIVSNSIPKREYYVATPEGRRLFDMSLGP
AALSFVGASGKDDLKRIRALRSEHGFDWPIHWLQTRGVHDAASLLNFE
MVALKRFRATGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAILSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKTTALSK
YIYSDEESRKKSYADTVLFIFKNAVREIEQYFANTLSVRRMETRETVERGGERIARYDELLQFVRFCITG
ESHPIRLPDVPMYLDWIATAELEHGLTPKIESRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQLFQTQSRSFDQDAMTMVTETEDAIAQASSQLVAYGYYTPVVVL
FDHDREALQEKAEAIRRLIQAEGFGARIETLNATDAFLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNTVAPCPFYPPNSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYESAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAGEGRALAFCPLSDLKSDADRAWATEWIEMLVGLQRVTITPDHRN
AISRQVGLMASAAGRSLSDFVSGVQMREIKDALHHYTVDGPMGLLLDAEEDGLSLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRSKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNVAAREPGTREFYERIGFNERQIEIVSNSIPKREYYVATPEGRRLFDMSLGP
AALSFVGASGKDDLKRIRALRSEHGFDWPIHWLQTRGVHDAASLLNFE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 257454..266833
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| N2599_RS35275 (N2599_35275) | 253139..254692 | - | 1554 | WP_027513949 | IS21 family transposase | - |
| N2599_RS35280 (N2599_35280) | 254889..256509 | + | 1621 | Protein_242 | Tn3 family transposase | - |
| N2599_RS35285 (N2599_35285) | 257454..258494 | - | 1041 | WP_027513742 | P-type DNA transfer ATPase VirB11 | virB11 |
| N2599_RS35290 (N2599_35290) | 258533..259642 | - | 1110 | WP_027513743 | type IV secretion system protein VirB10 | virB10 |
| N2599_RS35295 (N2599_35295) | 259639..260520 | - | 882 | WP_027513744 | P-type conjugative transfer protein VirB9 | virB9 |
| N2599_RS35300 (N2599_35300) | 260517..261230 | - | 714 | WP_027513745 | type IV secretion system protein VirB8 | virB8 |
| N2599_RS35305 (N2599_35305) | 261217..261384 | - | 168 | WP_084606758 | type IV secretion system lipoprotein VirB7 | - |
| N2599_RS35310 (N2599_35310) | 261418..262308 | - | 891 | WP_027513746 | type IV secretion system protein | virB6 |
| N2599_RS35315 (N2599_35315) | 262360..263040 | - | 681 | WP_027513747 | type IV secretion system protein VirB5 | traI |
| N2599_RS35320 (N2599_35320) | 263054..265423 | - | 2370 | WP_027513748 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| N2599_RS35325 (N2599_35325) | 265423..265749 | - | 327 | WP_027513749 | type IV secretion system protein VirB3 | virB3 |
| N2599_RS35330 (N2599_35330) | 265749..266111 | - | 363 | WP_027513750 | pilin major subunit VirB2 | virB2 |
| N2599_RS35335 (N2599_35335) | 266141..266833 | - | 693 | WP_027513751 | type IV secretion system lytic transglycosylase VirB1 | virB1 |
| N2599_RS35340 (N2599_35340) | 267209..267457 | + | 249 | Protein_254 | transposase | - |
| N2599_RS35345 (N2599_35345) | 267867..268337 | + | 471 | WP_027513752 | transposase | - |
| N2599_RS35350 (N2599_35350) | 268334..268618 | + | 285 | Protein_256 | IS66 family insertion sequence element accessory protein TnpB | - |
| N2599_RS35355 (N2599_35355) | 268672..269422 | - | 751 | Protein_257 | IS6 family transposase | - |
| N2599_RS35360 (N2599_35360) | 269428..270057 | + | 630 | Protein_258 | bifunctional folylpolyglutamate synthase/dihydrofolate synthase | - |
| N2599_RS35365 (N2599_35365) | 270106..270426 | - | 321 | WP_132568927 | thioredoxin | - |
| N2599_RS35370 (N2599_35370) | 270479..270847 | - | 369 | WP_027513883 | hypothetical protein | - |
Region 2: 455432..469712
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| N2599_RS36325 (N2599_36325) | 453702..454265 | + | 564 | WP_027513361 | conjugative transfer signal peptidase TraF | - |
| N2599_RS36330 (N2599_36330) | 454255..455415 | + | 1161 | WP_027513362 | conjugal transfer protein TraB | - |
| N2599_RS36335 (N2599_36335) | 455432..456052 | + | 621 | WP_027513363 | TraH family protein | virB1 |
| N2599_RS36340 (N2599_36340) | 456224..456598 | - | 375 | WP_027513364 | DUF5615 family PIN-like protein | - |
| N2599_RS36345 (N2599_36345) | 456595..457227 | - | 633 | WP_027513365 | DUF433 domain-containing protein | - |
| N2599_RS36350 (N2599_36350) | 457515..457907 | - | 393 | Protein_456 | adenylate cyclase | - |
| N2599_RS36355 (N2599_36355) | 458434..458646 | - | 213 | WP_027513367 | helix-turn-helix domain-containing protein | - |
| N2599_RS36360 (N2599_36360) | 458864..459187 | + | 324 | WP_027513368 | transcriptional repressor TraM | - |
| N2599_RS36365 (N2599_36365) | 459191..459895 | - | 705 | WP_027513369 | autoinducer binding domain-containing protein | - |
| N2599_RS36370 (N2599_36370) | 460208..461506 | - | 1299 | WP_027513370 | IncP-type conjugal transfer protein TrbI | virB10 |
| N2599_RS36375 (N2599_36375) | 461518..461958 | - | 441 | WP_027513371 | conjugal transfer protein TrbH | - |
| N2599_RS36380 (N2599_36380) | 461962..462774 | - | 813 | WP_027513372 | P-type conjugative transfer protein TrbG | virB9 |
| N2599_RS36385 (N2599_36385) | 462791..463453 | - | 663 | WP_027513373 | conjugal transfer protein TrbF | virB8 |
| N2599_RS36390 (N2599_36390) | 463474..464622 | - | 1149 | WP_245209322 | P-type conjugative transfer protein TrbL | virB6 |
| N2599_RS36395 (N2599_36395) | 464658..464831 | - | 174 | WP_311319516 | entry exclusion protein TrbK | - |
| N2599_RS36400 (N2599_36400) | 464828..465631 | - | 804 | WP_027513376 | P-type conjugative transfer protein TrbJ | virB5 |
| N2599_RS36405 (N2599_36405) | 465603..468059 | - | 2457 | WP_027513377 | conjugal transfer protein TrbE | virb4 |
| N2599_RS36410 (N2599_36410) | 468070..468369 | - | 300 | WP_027513378 | conjugal transfer protein TrbD | virB3 |
| N2599_RS36415 (N2599_36415) | 468362..468745 | - | 384 | WP_027513379 | TrbC/VirB2 family protein | virB2 |
| N2599_RS36420 (N2599_36420) | 468735..469712 | - | 978 | WP_027513380 | P-type conjugative transfer ATPase TrbB | virB11 |
| N2599_RS36425 (N2599_36425) | 469732..470346 | - | 615 | WP_027513381 | acyl-homoserine-lactone synthase TraI | - |
Host bacterium
| ID | 16221 | GenBank | NZ_CP104145 |
| Plasmid name | pWSM1592_2 | Incompatibility group | - |
| Plasmid size | 470679 bp | Coordinate of oriT [Strand] | 450266..450299 [+] |
| Host baterium | Rhizobium sullae strain WSM1592 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nodD, nifT, nifZ, nifB, fixX, fixC, fixB, fixA, nifW, nifS, nodC, nodB, nodA, a9174_31165, nodF, nodE, nodJ, nodI, nifH, nifD, nifK, nifE, nifN, nifX, mLTONO_5203 |
| Anti-CRISPR | - |