Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 115786 |
| Name | oriT_pSU277_1 |
| Organism | Sinorhizobium medicae strain SU277 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP104148 (999022..999055 [+], 34 nt) |
| oriT length | 34 nt |
| IRs (inverted repeats) | 20..25, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 34 nt
>oriT_pSU277_1
ATCCAAGGGCGCAATTATACGTCGCAGGGCGACG
ATCCAAGGGCGCAATTATACGTCGCAGGGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file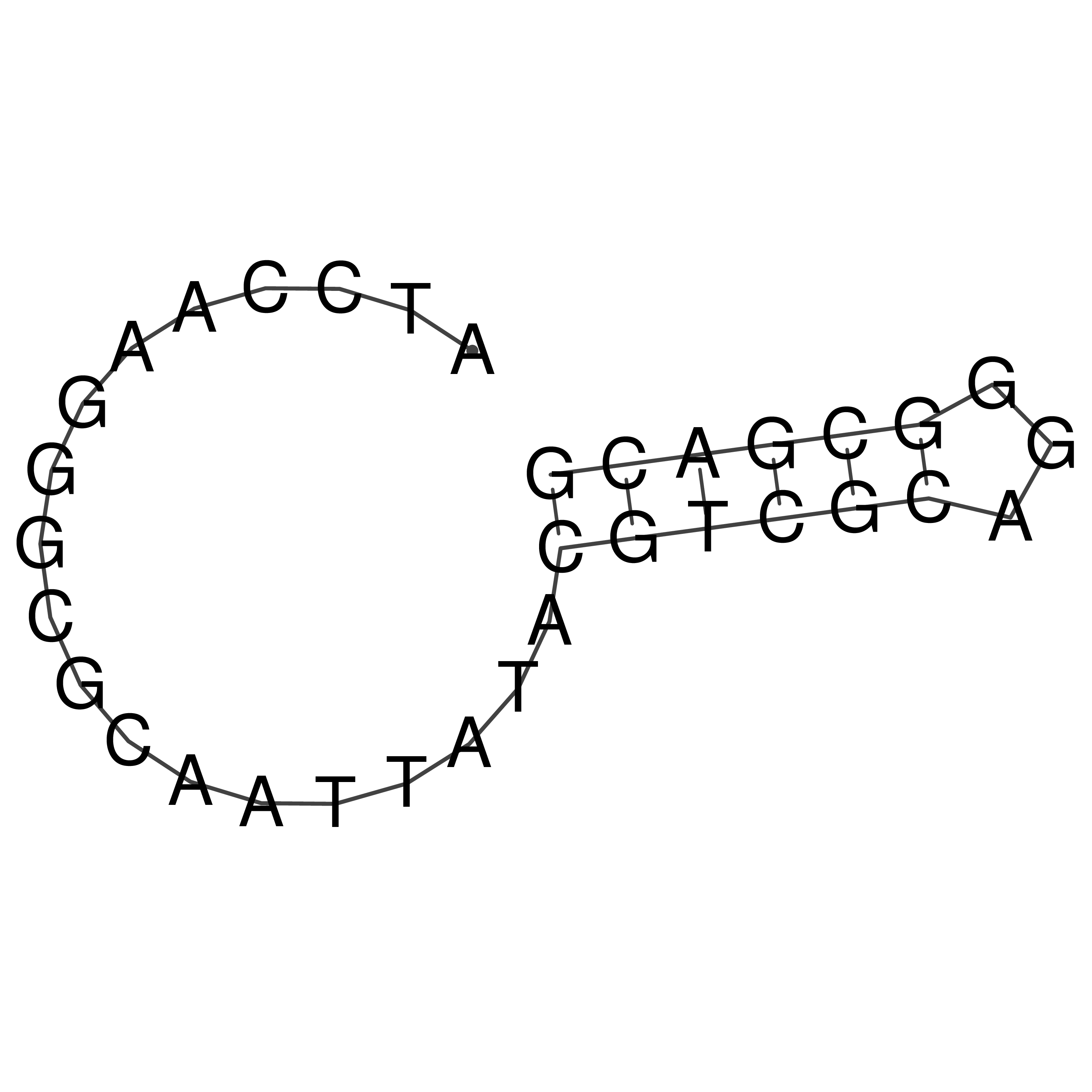
Relaxase
| ID | 10110 | GenBank | WP_260224799 |
| Name | traA_N2598_RS22555_pSU277_1 |
UniProt ID | _ |
| Length | 1102 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1102 a.a. Molecular weight: 122550.59 Da Isoelectric Point: 10.2256
>WP_260224799.1 Ti-type conjugative transfer relaxase TraA [Sinorhizobium medicae]
MAVPHFSVSLITRGSGRSAVLSAAYRHCAKMAYQREARTIDYTRKTGLLHEEFVIPDNAPEWLQSMITDH
SVSDAAEAFWNKVEDFEKRSDAQLAKEVTIALPIELTSAQNIALVRDFVEQHISAKGMVADWVYHDAPGN
PHVHLMTTLRPLTQQGFGAKKVAVLGPDGYPIRNDAGKIVYELWAGRLDDFNALRDGWFACQNRHLTLAG
LDIRIDGRSFEKQGIELAPTLHLGAGTKAIERKATAEAKTLSLERLQLQEDLRSENARRLQRRPEIVLDL
ITRERSVFDERDVAKILHRYVDDPAVFRSLMARILQSPETLQLERERIAFATGIRAPAKYTTRALIRLEA
EMANRAIWLSRQSSHRLREAVLEATLARHSRLSDEQKTAIEHVAKAGRIAAVIGRAGAGKTTMMKAAREA
WEAAGYRVVGGALAGKAAEGLEKEAGIISRTLSAWELRWSQGRNQLDARTVFILDEAGMVSSRQMACFIE
AATRTGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQREQWMRDASLDLARGNVGKAVEAYR
ANGRMIGAELKAEAVRNLIVDWNRDYDPTKTTLILAHLRRDVRMLNEMARSKLVERGIVGEGFAFRTADG
NRNFAAGDQIVFLKNEGSLGVKNGMLGKIVEASPNRVVAEIGAGEHHRQVIIEQRFYNNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLVYVAMTRHRDSLDVYYGARSFARAGGLIQVLSRKNSKETTLDYEKATFY
RQALRFAEARGLHLLNVARTIARDRLAWTARQSSRLADLAHRLVRIGRKLGLAAGTNPINSQNSKETRPM
VAGIATFPESVEQAVADKLAADPGLKKQWDDVSTRFHLVYAHPAAAFKAVNIDVMLKDETAAKATLSKIA
TQPDSYGALKGKTGLLASRADRQDRERAERNVPALVQSLDGYMRQRGSAERRYRAEESEARRKVAIDIPA
LSPGARQILERVRDAIDRNDLSAALEFARADRHVKAELDGFANAVDARFGKRTFLPLSARDTNGDTFTSV
TAGMNPGQRLEVESAWKAMRTVQQLSAHERTTEALKHSETLRLSKSQGLLLR
MAVPHFSVSLITRGSGRSAVLSAAYRHCAKMAYQREARTIDYTRKTGLLHEEFVIPDNAPEWLQSMITDH
SVSDAAEAFWNKVEDFEKRSDAQLAKEVTIALPIELTSAQNIALVRDFVEQHISAKGMVADWVYHDAPGN
PHVHLMTTLRPLTQQGFGAKKVAVLGPDGYPIRNDAGKIVYELWAGRLDDFNALRDGWFACQNRHLTLAG
LDIRIDGRSFEKQGIELAPTLHLGAGTKAIERKATAEAKTLSLERLQLQEDLRSENARRLQRRPEIVLDL
ITRERSVFDERDVAKILHRYVDDPAVFRSLMARILQSPETLQLERERIAFATGIRAPAKYTTRALIRLEA
EMANRAIWLSRQSSHRLREAVLEATLARHSRLSDEQKTAIEHVAKAGRIAAVIGRAGAGKTTMMKAAREA
WEAAGYRVVGGALAGKAAEGLEKEAGIISRTLSAWELRWSQGRNQLDARTVFILDEAGMVSSRQMACFIE
AATRTGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQREQWMRDASLDLARGNVGKAVEAYR
ANGRMIGAELKAEAVRNLIVDWNRDYDPTKTTLILAHLRRDVRMLNEMARSKLVERGIVGEGFAFRTADG
NRNFAAGDQIVFLKNEGSLGVKNGMLGKIVEASPNRVVAEIGAGEHHRQVIIEQRFYNNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLVYVAMTRHRDSLDVYYGARSFARAGGLIQVLSRKNSKETTLDYEKATFY
RQALRFAEARGLHLLNVARTIARDRLAWTARQSSRLADLAHRLVRIGRKLGLAAGTNPINSQNSKETRPM
VAGIATFPESVEQAVADKLAADPGLKKQWDDVSTRFHLVYAHPAAAFKAVNIDVMLKDETAAKATLSKIA
TQPDSYGALKGKTGLLASRADRQDRERAERNVPALVQSLDGYMRQRGSAERRYRAEESEARRKVAIDIPA
LSPGARQILERVRDAIDRNDLSAALEFARADRHVKAELDGFANAVDARFGKRTFLPLSARDTNGDTFTSV
TAGMNPGQRLEVESAWKAMRTVQQLSAHERTTEALKHSETLRLSKSQGLLLR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 11813 | GenBank | WP_018210029 |
| Name | tcpA_N2598_RS18485_pSU277_1 |
UniProt ID | _ |
| Length | 951 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 951 a.a. Molecular weight: 102047.57 Da Isoelectric Point: 4.5063
>WP_018210029.1 DNA translocase FtsK [Sinorhizobium medicae]
MRIPRTNFSTAALYDANQAEDFEDPQPVVPGIVHASAPESAAHDHLFEAPEAPRRASGYRQEEIGQATRA
VRLYGHGSAYAPAEVTVSTRDPLPTLAEITGLSCEPGWESHFFLSPNVRFTRTPEREFMKRRPPVPEDNE
VDAAEAAAEAPLAETVTGKEPVEPPPSPAETDAPSYSPSELLRVLVQQLPSFNAAHLQTPEKNAVEQAAE
AAILTEEEPSIPQAPHVPIMEEAPVVADASTGTAAVPDSAGAEDVARQAEARLSYLSDFAFFEFMPLEPP
VALRTVAAPAEESAHVPAPSAALPKAAAPNIVSAPVRIPSQPAAAITSLFRVVECRRPEAGPDVSAPDAP
QDLGAEPISAGAAVGAEAVEAQGLTPEAPDEPAREPAPEAAVVPAKEPAPEAPVTRAAITMPAVIQRSSP
ALPPVGATERPGIADAYEFPSKELLQEPPQGQGFFMTQEQLEQNAGLLESVLEDFGVKGEIIHVRPGPVV
TLYEFEPAPGVKSSRVIGLADDIARSMSALSARVAVVPGRNVIGIELPNATRETVYFRELIESGDFQKTG
CKLALCLGKTIGGEPVIAELAKMPHLLVAGTTGSGKSVAINTMILSLLYRLKPEECRLIMVDPKMLELSV
YDGIPHLLTPVVTDPKKAVMALKWAVREMEDRYRKMSRLGVRNIDGYNQRAAAAREKGEPILATVQTGFE
KGTGEPLFEQQEMDLSPMPYIVVIVDEMADLMMVAGKEIEGAIQRLAQMARAAGIHLIMATQRPSVDVIT
GTIKANFPTRISFQVTSKIDSRTILGEQGAEQLLGQGDMLHMAGGGRIARVHGPFVSDQEVEHVVAHLKT
QGRPEYLETVTADEEEEEPEEDQGAVFDKSAIAVEDGNELYDQAVKVVLRDKKCSTSYIQRRLGIGYNRA
ASLVERMEKDGLVGPANHVGKREIIYGNRDGGPKPDSDELD
MRIPRTNFSTAALYDANQAEDFEDPQPVVPGIVHASAPESAAHDHLFEAPEAPRRASGYRQEEIGQATRA
VRLYGHGSAYAPAEVTVSTRDPLPTLAEITGLSCEPGWESHFFLSPNVRFTRTPEREFMKRRPPVPEDNE
VDAAEAAAEAPLAETVTGKEPVEPPPSPAETDAPSYSPSELLRVLVQQLPSFNAAHLQTPEKNAVEQAAE
AAILTEEEPSIPQAPHVPIMEEAPVVADASTGTAAVPDSAGAEDVARQAEARLSYLSDFAFFEFMPLEPP
VALRTVAAPAEESAHVPAPSAALPKAAAPNIVSAPVRIPSQPAAAITSLFRVVECRRPEAGPDVSAPDAP
QDLGAEPISAGAAVGAEAVEAQGLTPEAPDEPAREPAPEAAVVPAKEPAPEAPVTRAAITMPAVIQRSSP
ALPPVGATERPGIADAYEFPSKELLQEPPQGQGFFMTQEQLEQNAGLLESVLEDFGVKGEIIHVRPGPVV
TLYEFEPAPGVKSSRVIGLADDIARSMSALSARVAVVPGRNVIGIELPNATRETVYFRELIESGDFQKTG
CKLALCLGKTIGGEPVIAELAKMPHLLVAGTTGSGKSVAINTMILSLLYRLKPEECRLIMVDPKMLELSV
YDGIPHLLTPVVTDPKKAVMALKWAVREMEDRYRKMSRLGVRNIDGYNQRAAAAREKGEPILATVQTGFE
KGTGEPLFEQQEMDLSPMPYIVVIVDEMADLMMVAGKEIEGAIQRLAQMARAAGIHLIMATQRPSVDVIT
GTIKANFPTRISFQVTSKIDSRTILGEQGAEQLLGQGDMLHMAGGGRIARVHGPFVSDQEVEHVVAHLKT
QGRPEYLETVTADEEEEEPEEDQGAVFDKSAIAVEDGNELYDQAVKVVLRDKKCSTSYIQRRLGIGYNRA
ASLVERMEKDGLVGPANHVGKREIIYGNRDGGPKPDSDELD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 16219 | GenBank | NZ_CP104148 |
| Plasmid name | pSU277_1 | Incompatibility group | - |
| Plasmid size | 1534656 bp | Coordinate of oriT [Strand] | 999022..999055 [+] |
| Host baterium | Sinorhizobium medicae strain SU277 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | actP |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA9, AcrIIA7 |