Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 115782 |
| Name | oriT_pWSM1274_1 |
| Organism | Rhizobium sp. WSM1274 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP104139 (882331..882359 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pWSM1274_1
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file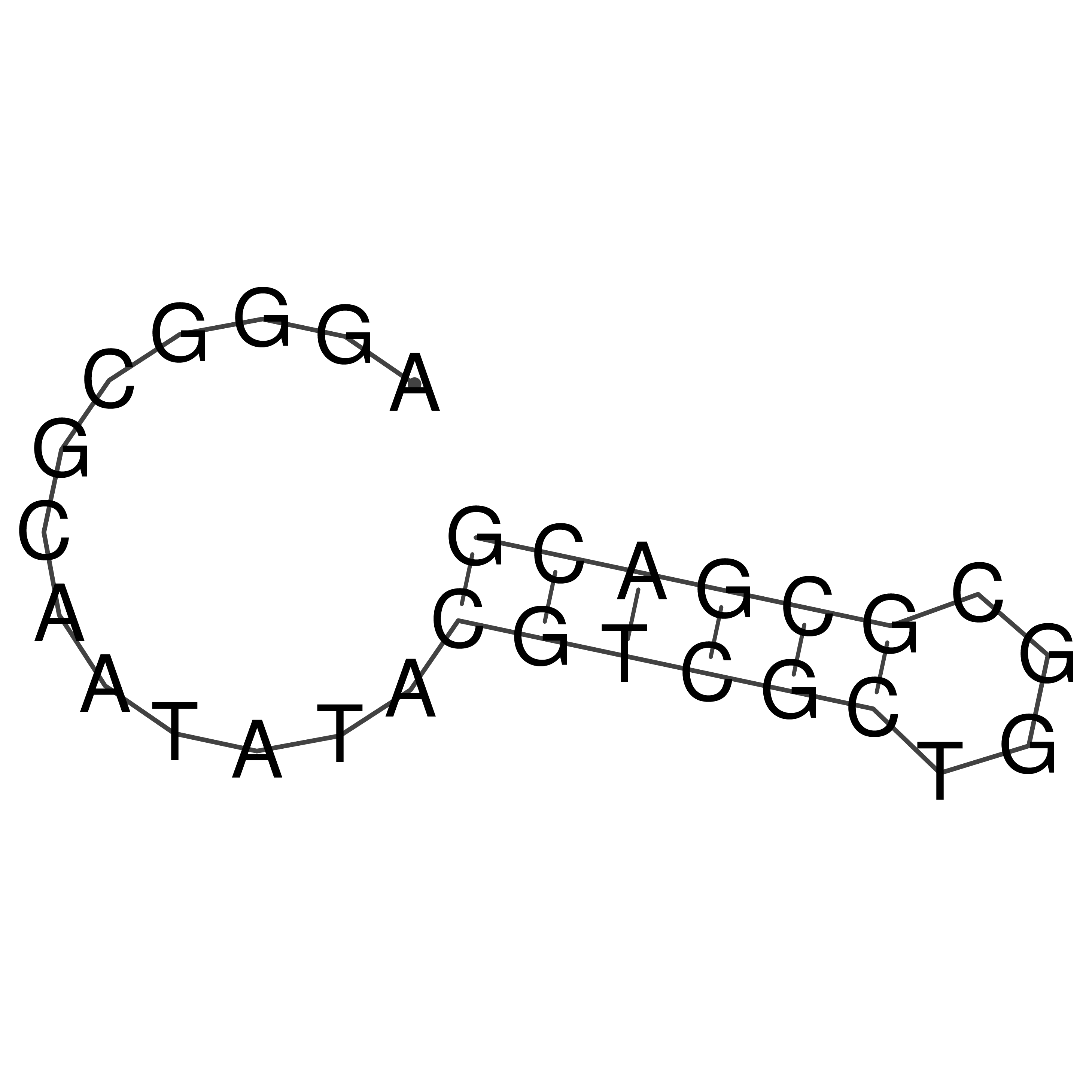
Relaxase
| ID | 10106 | GenBank | WP_260299030 |
| Name | traA_N2600_RS28435_pWSM1274_1 |
UniProt ID | _ |
| Length | 1543 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1543 a.a. Molecular weight: 171194.26 Da Isoelectric Point: 6.7948
>WP_260299030.1 Ti-type conjugative transfer relaxase TraA [Rhizobium sp. WSM1274]
MAIMFVRAQVIGRGGGRSIVSAAAYRHRTRMMDEQAGTSFSYRGGASELVHEELALPDQTPDWLRAAIDG
RTVAAASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVFHDKDG
NPHIHLMTTLRPLTEEGFGRKKVPVTGEDGTPLRVVTPDRPNGRIVYKLWAGDKETMKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGRELYFAPADLARRQEMADRLLAEPELLLKQ
LGNERSTFDERDIAKALHRYVDDPLVFANIRARLMASDDLIMLKPQQIDAETGKASEPAVFTTREILRIE
YDMARSAQVLSERRGFAVSSRHVAAAVRHVETGDPEKPFKLDAEQVDAIRHVTGDNGIAAVVGLAGAGKS
TLLAGARVAWEHDGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELIWANGHETLHRGDVLVIDEAGMVS
SQQMARVLKIAEEAGTKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRDDWAREASRLFARGE
VEKGLDAYAQQGHLVEAATRDEIIGRIVSDWTDARRALVRETAEGANTTRLRGDELLVLAHTNQDVKRLN
EALRLVMTDEGALGESRSFRTERGAREFAAGDRIIFLENARFLEPRAPRLGPQYVKNGMLGTVVSTGDKR
GDTLLSVRLDNGRDVVISENSYRNVDHGYAATIHKSQGVTVERTFVLATGMMDQHLTYVSMTRHRDRADL
YAAKDDFEARPEWGRKPRVDHAAGVTGALVETGQAKFRPNDEDADDSPYADVKADDGTLHRLWGVSLPKA
LEEGGVSQGDTVTLRKDGVEKVKVQVQIVDEQTGQKRFEEKEVDRNVWTAKQIETAEARLERIERESHRP
EVFKQLVLRLSRSGAKTTTLDFESEAGYRAHADDFARRRGLDHLSQAAAGMEESLSRQWAWIAEKREQVA
ELWERASMALGFAIERERRVSYNEERIETQTVSDADADRARHLIAPTTVFARSVEEDARLAQLSSPAWKE
REAILRPLLEKIYRDPDAALSALNALSSDMSIEPRRLADDLAAAPDRLGRLRGSDLIVDGRAARDERNAA
ITALTDLLPLARAHATEFRRNAERFESREQQRRTHMSLPIPALSKTAMTRLAEIEAVRRQGGDDAYKTAF
ALAAEDRSFVQEVKAVSEALSARFGWSAFTSKTDAVAERNMIERMPEDLASISGEKLIRLFEAVKRFADE
QHQVERRDRSKIVAAASADQGMEADKENAAVLPMLAAITEFKNPLDEEARSRLLSDPLYRQQRAALAAVA
STIWRDPADAVGKIEDLLAKGFAGERIAAAVTNDPAAYGALRGSDRLMDRMLAAGRERKEAVQAVPEAAA
RLRALGSAYVNVLDAERQAVAEERRRMAVAIPGLSKAAEEALMRLTAEARNNGRKLNASAASLDPDIHRE
FAAVSRALDERFGRNAIIRGEKDLIHRVPQMQRNVFEAMQEKLKVLQQAVRRESSEQIISERRQRAVSRG
REI
MAIMFVRAQVIGRGGGRSIVSAAAYRHRTRMMDEQAGTSFSYRGGASELVHEELALPDQTPDWLRAAIDG
RTVAAASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVFHDKDG
NPHIHLMTTLRPLTEEGFGRKKVPVTGEDGTPLRVVTPDRPNGRIVYKLWAGDKETMKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGRELYFAPADLARRQEMADRLLAEPELLLKQ
LGNERSTFDERDIAKALHRYVDDPLVFANIRARLMASDDLIMLKPQQIDAETGKASEPAVFTTREILRIE
YDMARSAQVLSERRGFAVSSRHVAAAVRHVETGDPEKPFKLDAEQVDAIRHVTGDNGIAAVVGLAGAGKS
TLLAGARVAWEHDGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELIWANGHETLHRGDVLVIDEAGMVS
SQQMARVLKIAEEAGTKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRDDWAREASRLFARGE
VEKGLDAYAQQGHLVEAATRDEIIGRIVSDWTDARRALVRETAEGANTTRLRGDELLVLAHTNQDVKRLN
EALRLVMTDEGALGESRSFRTERGAREFAAGDRIIFLENARFLEPRAPRLGPQYVKNGMLGTVVSTGDKR
GDTLLSVRLDNGRDVVISENSYRNVDHGYAATIHKSQGVTVERTFVLATGMMDQHLTYVSMTRHRDRADL
YAAKDDFEARPEWGRKPRVDHAAGVTGALVETGQAKFRPNDEDADDSPYADVKADDGTLHRLWGVSLPKA
LEEGGVSQGDTVTLRKDGVEKVKVQVQIVDEQTGQKRFEEKEVDRNVWTAKQIETAEARLERIERESHRP
EVFKQLVLRLSRSGAKTTTLDFESEAGYRAHADDFARRRGLDHLSQAAAGMEESLSRQWAWIAEKREQVA
ELWERASMALGFAIERERRVSYNEERIETQTVSDADADRARHLIAPTTVFARSVEEDARLAQLSSPAWKE
REAILRPLLEKIYRDPDAALSALNALSSDMSIEPRRLADDLAAAPDRLGRLRGSDLIVDGRAARDERNAA
ITALTDLLPLARAHATEFRRNAERFESREQQRRTHMSLPIPALSKTAMTRLAEIEAVRRQGGDDAYKTAF
ALAAEDRSFVQEVKAVSEALSARFGWSAFTSKTDAVAERNMIERMPEDLASISGEKLIRLFEAVKRFADE
QHQVERRDRSKIVAAASADQGMEADKENAAVLPMLAAITEFKNPLDEEARSRLLSDPLYRQQRAALAAVA
STIWRDPADAVGKIEDLLAKGFAGERIAAAVTNDPAAYGALRGSDRLMDRMLAAGRERKEAVQAVPEAAA
RLRALGSAYVNVLDAERQAVAEERRRMAVAIPGLSKAAEEALMRLTAEARNNGRKLNASAASLDPDIHRE
FAAVSRALDERFGRNAIIRGEKDLIHRVPQMQRNVFEAMQEKLKVLQQAVRRESSEQIISERRQRAVSRG
REI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 11808 | GenBank | WP_260299029 |
| Name | traG_N2600_RS28420_pWSM1274_1 |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70472.34 Da Isoelectric Point: 8.6943
>WP_260299029.1 Ti-type conjugative transfer system protein TraG [Rhizobium sp. WSM1274]
MALKCRLHPGLLLVLVPVAVTAIAVYITGWRWPALAHGLSGKSEYWFMRAAPVPALLFGPLAGLLIVWAL
PLHRRRSIAVASLLCFLVMSSFYVLREYGRLAPSVESGTVSWDRALSYLDFVAVIGALAGFMTVAVAARI
SVVVPEQVRRAKGGTFGDADWLSMSAVAKLFPPDGEIVVGERYRVDKEIVHELPFDPNDRNTWGQGGKAP
LLTYSQDFDSTHMLFFAGSGGYKTTSNVLPTALRYTGPLICLDPSTEVAPMVLEHRTRTLGREVMVLDPT
NPIMGFNVLDGIEASKQKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSESAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFRSSDIVAGGTDVFLNIPASILRSYPGIGRVIIGSLINAMIQADGAFSRRALFMLDEVDLLGYMRIL
EEARDRGRKYGITMMLMYQSVGQMERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVQG
RSRNIGWDTKNGGTRKSESVNFQRRPLIMPHEITQSMRKDEQIIIVQGHPPIRCGRAIYFRRKDMNAAAK
ANRFVKPGP
MALKCRLHPGLLLVLVPVAVTAIAVYITGWRWPALAHGLSGKSEYWFMRAAPVPALLFGPLAGLLIVWAL
PLHRRRSIAVASLLCFLVMSSFYVLREYGRLAPSVESGTVSWDRALSYLDFVAVIGALAGFMTVAVAARI
SVVVPEQVRRAKGGTFGDADWLSMSAVAKLFPPDGEIVVGERYRVDKEIVHELPFDPNDRNTWGQGGKAP
LLTYSQDFDSTHMLFFAGSGGYKTTSNVLPTALRYTGPLICLDPSTEVAPMVLEHRTRTLGREVMVLDPT
NPIMGFNVLDGIEASKQKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSESAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFRSSDIVAGGTDVFLNIPASILRSYPGIGRVIIGSLINAMIQADGAFSRRALFMLDEVDLLGYMRIL
EEARDRGRKYGITMMLMYQSVGQMERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVQG
RSRNIGWDTKNGGTRKSESVNFQRRPLIMPHEITQSMRKDEQIIIVQGHPPIRCGRAIYFRRKDMNAAAK
ANRFVKPGP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 365984..375585
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| N2600_RS25945 (N2600_25945) | 361494..362069 | - | 576 | WP_260299315 | TadE/TadG family type IV pilus assembly protein | - |
| N2600_RS25950 (N2600_25950) | 362071..362601 | - | 531 | WP_222281789 | TadE/TadG family type IV pilus assembly protein | - |
| N2600_RS25955 (N2600_25955) | 362598..363959 | - | 1362 | WP_260299485 | TadE/TadG family type IV pilus assembly protein | - |
| N2600_RS25960 (N2600_25960) | 364996..365349 | - | 354 | WP_170281502 | MarR family transcriptional regulator | - |
| N2600_RS25965 (N2600_25965) | 365441..365980 | + | 540 | WP_260299316 | hypothetical protein | - |
| N2600_RS25970 (N2600_25970) | 365984..366646 | + | 663 | WP_260299317 | lytic transglycosylase domain-containing protein | virB1 |
| N2600_RS25975 (N2600_25975) | 366643..366942 | + | 300 | WP_222281786 | TrbC/VirB2 family protein | virB2 |
| N2600_RS25980 (N2600_25980) | 366947..367285 | + | 339 | WP_260299318 | type IV secretion system protein VirB3 | virB3 |
| N2600_RS25985 (N2600_25985) | 367278..369644 | + | 2367 | WP_260299319 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| N2600_RS25990 (N2600_25990) | 369677..370342 | + | 666 | WP_260299486 | P-type DNA transfer protein VirB5 | virB5 |
| N2600_RS25995 (N2600_25995) | 370339..370572 | + | 234 | WP_222387198 | EexN family lipoprotein | - |
| N2600_RS26000 (N2600_26000) | 370576..371496 | + | 921 | WP_260299320 | type IV secretion system protein | virB6 |
| N2600_RS26005 (N2600_26005) | 371550..371834 | + | 285 | WP_260299321 | hypothetical protein | - |
| N2600_RS26010 (N2600_26010) | 371836..372507 | + | 672 | WP_260299322 | virB8 family protein | virB8 |
| N2600_RS26015 (N2600_26015) | 372504..373358 | + | 855 | WP_260299323 | P-type conjugative transfer protein VirB9 | virB9 |
| N2600_RS26020 (N2600_26020) | 373368..374537 | + | 1170 | WP_260299324 | type IV secretion system protein VirB10 | virB10 |
| N2600_RS26025 (N2600_26025) | 374545..375585 | + | 1041 | WP_260299325 | P-type DNA transfer ATPase VirB11 | virB11 |
| N2600_RS26030 (N2600_26030) | 375743..375958 | - | 216 | WP_260299326 | DUF3072 domain-containing protein | - |
| N2600_RS26035 (N2600_26035) | 376302..377210 | - | 909 | WP_222387190 | nucleotidyltransferase and HEPN domain-containing protein | - |
| N2600_RS26040 (N2600_26040) | 377311..377604 | - | 294 | Protein_363 | integrase | - |
| N2600_RS26045 (N2600_26045) | 377621..378007 | - | 387 | WP_260299327 | type II toxin-antitoxin system VapC family toxin | - |
| N2600_RS26050 (N2600_26050) | 378004..378252 | - | 249 | WP_162118950 | type II toxin-antitoxin system VapB family antitoxin | - |
| N2600_RS26055 (N2600_26055) | 378429..379222 | - | 794 | Protein_366 | tyrosine-type recombinase/integrase | - |
| N2600_RS26060 (N2600_26060) | 379410..380549 | + | 1140 | WP_260299487 | RHE_PE00001 family protein | - |
Host bacterium
| ID | 16215 | GenBank | NZ_CP104139 |
| Plasmid name | pWSM1274_1 | Incompatibility group | - |
| Plasmid size | 1180112 bp | Coordinate of oriT [Strand] | 882331..882359 [+] |
| Host baterium | Rhizobium sp. WSM1274 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | fixA, fixB, nifN, nifE, nifK, nifD, nifH, nodM, nodE, nodF, nodD, nodA, nodC, nodI, nodJ, nifB, fixX, fixC, fixN, fixO, fixQ, fixG |
| Anti-CRISPR | - |