Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 115579 |
| Name | oriT_pFB-5 |
| Organism | Sphaerotilus microaerophilus strain FB-5 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_AP025731 (24884..24980 [-], 97 nt) |
| oriT length | 97 nt |
| IRs (inverted repeats) | 24..31, 34..41 (TAACCGGC..GCCGGTTA) |
| Location of nic site | 60..61 |
| Conserved sequence flanking the nic site |
TCCTGCCCGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 97 nt
>oriT_pFB-5
GAATAAGGGACAGCGAAGATAGATAACCGGCCCGCCGGTTAGCTAACTTCGCACATCCTGCCCGCCTTACGGCGTTGATAACACCAAGGAAAGTCTA
GAATAAGGGACAGCGAAGATAGATAACCGGCCCGCCGGTTAGCTAACTTCGCACATCCTGCCCGCCTTACGGCGTTGATAACACCAAGGAAAGTCTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file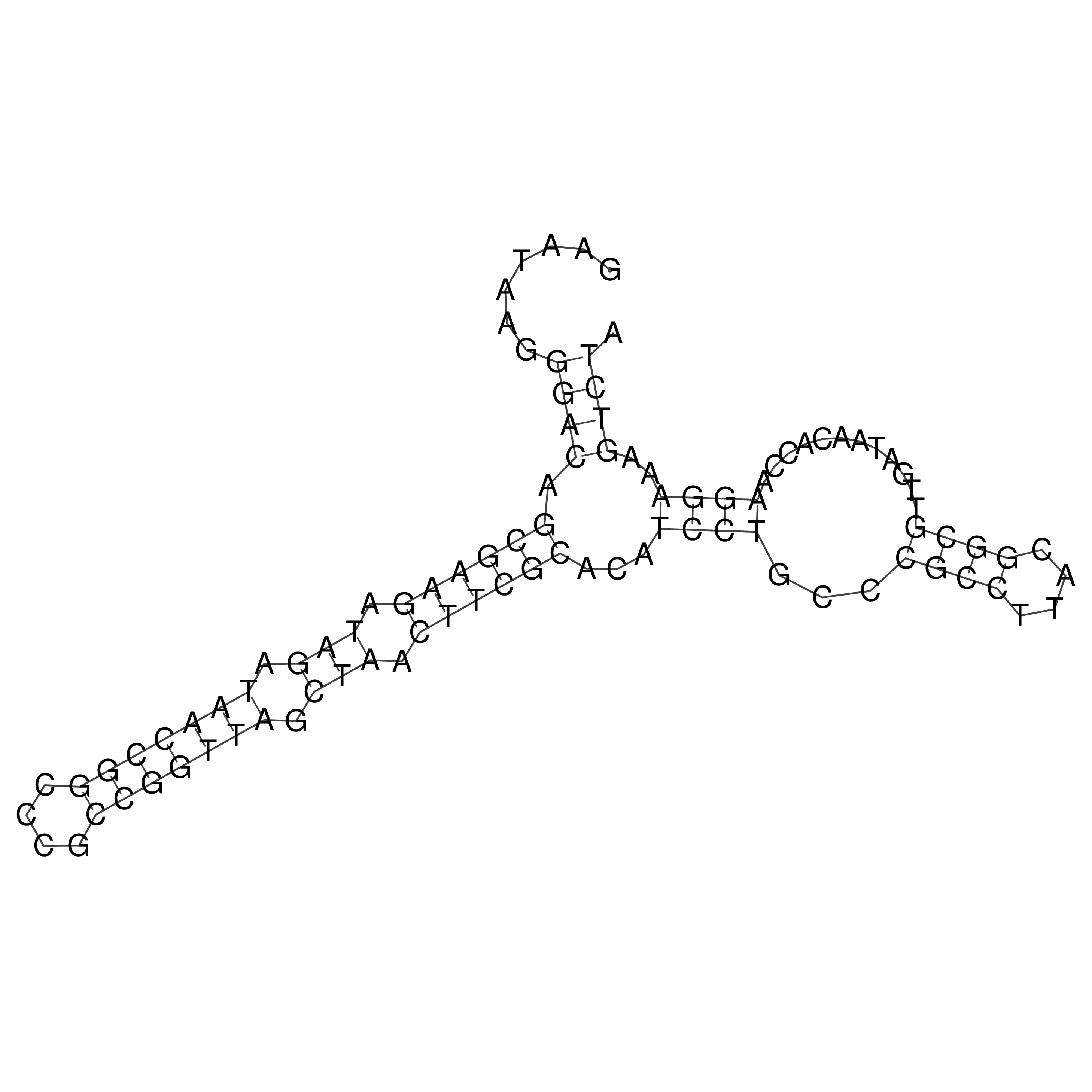
Relaxase
| ID | 9973 | GenBank | WP_012478238 |
| Name | traI_NGK70_RS26345_pFB-5 |
UniProt ID | A0A1H7CDK8 |
| Length | 752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 752 a.a. Molecular weight: 82867.55 Da Isoelectric Point: 10.6815
>WP_012478238.1 MULTISPECIES: TraI/MobA(P) family conjugative relaxase [Pseudomonadota]
MIAKHVPMRSLGKSDFAGLVNYVTDAQSKDHRLGQVQITNCDAVSVQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICAGLGYGEHQRVSAVHNDTDNLHIHIAINKIHPTRNTMHEPYYSHRA
LAELCTALERDYGLERDNHEPRQRGAEGRAADMEQHAGVESLVGWIKRECLDEIKGAQSWQELHQVMRDN
GLELRERANGLVIEAGDGTMVKASTLARDLSKPKLEARLGPFEASPERQTRTKAKRQYQKDPIRLRVNTV
ELYARYKAEQQTLTATRGQALEQARRRKDRLVEAAKRSGRLRRATIKVIGESGPNKKLLYAQASKALRDE
IQAINKQYQQDRQALYDSHSRRTWADWLKKEATHGNAEALTALRAREAAQGLKGNTIQGRGDPRPGHAPV
TDNITKKGTIIFRAGLTAVRDDGDKLQVSREATREGLQEALRLAMERYGSRITVNGTTQFQAQIIRAAVD
SQLPITFADPALESRRLALLKKENTHERPDRTERADQRPAEHRGRAGRGPGGPGQRAATHDDAARPIAVA
RAGFGRPGGGRAGTAGVHAGAVHRKPDIGRIGRVPPPQSQHRLRTLSQLGVVRIAGGSEVLLPRDVPRHL
EQQGTQPDHQLRRDISRPGSGVAAAPPGLAAADKYIAERESKRSKGFDIPKHARYTAGEGGLAFEGVRNI
EGQALALLKRGDEVMVMPIDQATARRMSRIAVGDAVTITPKGSIKTSKGRSR
MIAKHVPMRSLGKSDFAGLVNYVTDAQSKDHRLGQVQITNCDAVSVQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICAGLGYGEHQRVSAVHNDTDNLHIHIAINKIHPTRNTMHEPYYSHRA
LAELCTALERDYGLERDNHEPRQRGAEGRAADMEQHAGVESLVGWIKRECLDEIKGAQSWQELHQVMRDN
GLELRERANGLVIEAGDGTMVKASTLARDLSKPKLEARLGPFEASPERQTRTKAKRQYQKDPIRLRVNTV
ELYARYKAEQQTLTATRGQALEQARRRKDRLVEAAKRSGRLRRATIKVIGESGPNKKLLYAQASKALRDE
IQAINKQYQQDRQALYDSHSRRTWADWLKKEATHGNAEALTALRAREAAQGLKGNTIQGRGDPRPGHAPV
TDNITKKGTIIFRAGLTAVRDDGDKLQVSREATREGLQEALRLAMERYGSRITVNGTTQFQAQIIRAAVD
SQLPITFADPALESRRLALLKKENTHERPDRTERADQRPAEHRGRAGRGPGGPGQRAATHDDAARPIAVA
RAGFGRPGGGRAGTAGVHAGAVHRKPDIGRIGRVPPPQSQHRLRTLSQLGVVRIAGGSEVLLPRDVPRHL
EQQGTQPDHQLRRDISRPGSGVAAAPPGLAAADKYIAERESKRSKGFDIPKHARYTAGEGGLAFEGVRNI
EGQALALLKRGDEVMVMPIDQATARRMSRIAVGDAVTITPKGSIKTSKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A1H7CDK8 |
Auxiliary protein
| ID | 5850 | GenBank | WP_011255188 |
| Name | WP_011255188_pFB-5 |
UniProt ID | A0A1H7CD62 |
| Length | 123 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 123 a.a. Molecular weight: 13964.97 Da Isoelectric Point: 8.4531
>WP_011255188.1 MULTISPECIES: conjugal transfer transcriptional regulator TraJ [Pseudomonadota]
MEADEQPTAKRRQHLRVPVFPDEKEQIEANAKRAGVSVARYLRDVGQGYQIRGVMDYEYVRELVRVNGDL
GRLGGLLKLWLTDDPRTARFGDATILALLGRIEATQDEMSRLMKSVVQPRAEP
MEADEQPTAKRRQHLRVPVFPDEKEQIEANAKRAGVSVARYLRDVGQGYQIRGVMDYEYVRELVRVNGDL
GRLGGLLKLWLTDDPRTARFGDATILALLGRIEATQDEMSRLMKSVVQPRAEP
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A1H7CD62 |
T4CP
| ID | 11632 | GenBank | WP_012478237 |
| Name | t4cp2_NGK70_RS26350_pFB-5 |
UniProt ID | _ |
| Length | 634 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 634 a.a. Molecular weight: 69616.60 Da Isoelectric Point: 8.4953
>WP_012478237.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Pseudomonadota]
MKIKMNNAVGPQVRSAKPKPSKLLPILGGVSLVGGLQAATQFFAYTFQYHASLGANLGHVYAPWSILNWS
AKWYSQYPDEIMRAGSIGMVTATVGLLGVAVAKVVTSNSSKANEYLHGSARWAEKKDIQAAGLLPRERNV
LEIVTGKDAPTATGVYVGGWQDKDGNFYYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASSSGGVCWNPLDEIRVGTEAEVGDVQNLATLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKARNDGGTATLPSVDAMLADPNRDIGELWMEMTTYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLALYRDPVVARSVSRSDFRIKDLMHSDDPVSLYIVTQPNDKARLRPLVRVMVN
MIVRLLADKMDFENGRPVAHYKHRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLRSRET
GYGHDESITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTYQEVQRPLLT
PDECLRMPGPKKNDKGEIEEAGDMVIYVAGYPAIYGKQPLYFKDPVFQARASIPAPKATDRLRQVVEAGE
GITI
MKIKMNNAVGPQVRSAKPKPSKLLPILGGVSLVGGLQAATQFFAYTFQYHASLGANLGHVYAPWSILNWS
AKWYSQYPDEIMRAGSIGMVTATVGLLGVAVAKVVTSNSSKANEYLHGSARWAEKKDIQAAGLLPRERNV
LEIVTGKDAPTATGVYVGGWQDKDGNFYYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASSSGGVCWNPLDEIRVGTEAEVGDVQNLATLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKARNDGGTATLPSVDAMLADPNRDIGELWMEMTTYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLALYRDPVVARSVSRSDFRIKDLMHSDDPVSLYIVTQPNDKARLRPLVRVMVN
MIVRLLADKMDFENGRPVAHYKHRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLRSRET
GYGHDESITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTYQEVQRPLLT
PDECLRMPGPKKNDKGEIEEAGDMVIYVAGYPAIYGKQPLYFKDPVFQARASIPAPKATDRLRQVVEAGE
GITI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 16012 | GenBank | NZ_AP025731 |
| Plasmid name | pFB-5 | Incompatibility group | - |
| Plasmid size | 30838 bp | Coordinate of oriT [Strand] | 24884..24980 [-] |
| Host baterium | Sphaerotilus microaerophilus strain FB-5 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |