Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 115430 |
| Name | oriT_p32 |
| Organism | Enterococcus faecium strain 515 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP041255 (40063..40255 [+], 193 nt) |
| oriT length | 193 nt |
| IRs (inverted repeats) | 123..128, 142..147 (AAAATA..TATTTT) 123..128, 131..136 (AAAATA..TATTTT) 104..111, 122..129 (ATATTTTT..AAAAATAT) 109..114, 121..126 (TTTTTG..CAAAAA) 65..71, 75..81 (AGTTGGC..GCCAACT) 41..48, 53..60 (CACCTTCC..GGAAGGTG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 193 nt
>oriT_p32
CCAAGGAATTAATACAAAGAGCATAAGGGAAAACTGATAGCACCTTCCCAAAGGAAGGTGGCTAAGTTGGCTGTGCCAACTGGTTTTCTTTCAAAATCACCTCATATTTTTTTGCTATTACAAAAATATCTATTTTCGACCTATTTTCGGCCATAATATAGTACCTACTTTTGGTCATAGTTTCGTCCGTAGT
CCAAGGAATTAATACAAAGAGCATAAGGGAAAACTGATAGCACCTTCCCAAAGGAAGGTGGCTAAGTTGGCTGTGCCAACTGGTTTTCTTTCAAAATCACCTCATATTTTTTTGCTATTACAAAAATATCTATTTTCGACCTATTTTCGGCCATAATATAGTACCTACTTTTGGTCATAGTTTCGTCCGTAGT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file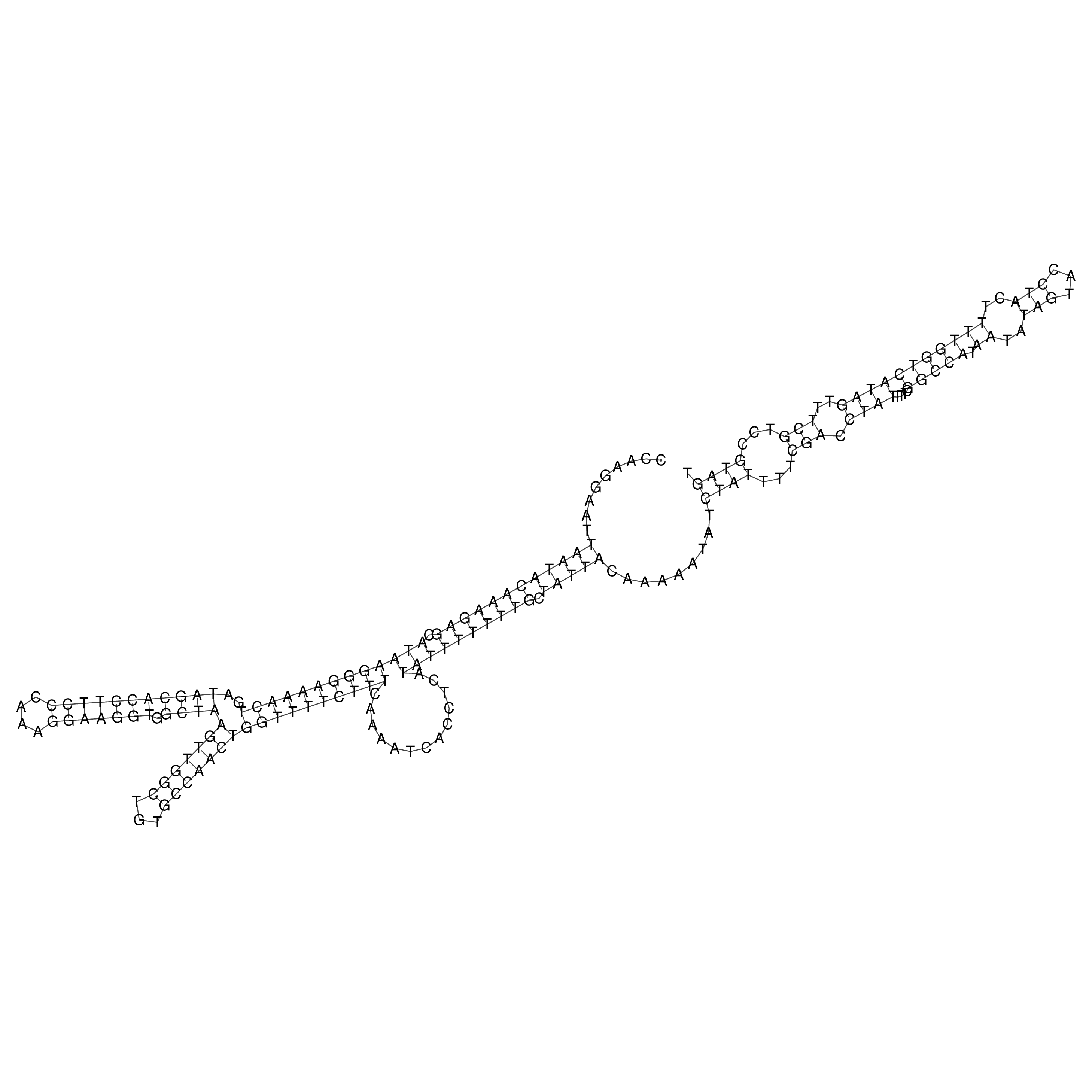
Relaxase
| ID | 9876 | GenBank | WP_002326870 |
| Name | mobP2_FKZ20_RS00625_p32 |
UniProt ID | B5U8V6 |
| Length | 506 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 506 a.a. Molecular weight: 60597.01 Da Isoelectric Point: 9.3485
>WP_002326870.1 MULTISPECIES: MobP2 family relaxase [Enterococcus]
MTEHIFNRKSASIILKAKFETGKSKKNNHMVELQKFVDYISRQEAIRQDKLDHDYSEDELNELARIEKAL
EKIEQETDQPVIKDLREMDKYIDYMTRKKAILENVDNEIVNGAFSNTKRYITKKDISKIKESVIEAKNNG
SVMFQDVISFDNDFLVREGYYNPETNELNENVLYEATKSMMGKMQEKEELVDPFWFATIHRNTEHIHIHV
TAMERKNTREIMEYDGVLQARGKRKQSTLDDMIFKFGSKILDRTNEFEKISKLRKEVPLELKQSVKDSLI
QLYVENNSRYDKELVKYLKELKKEIPSTTRGYNELPEETKEKIDEVTNYMTKDNPKRKEYDRMTKEIDEL
YQTTYGKRYNPQEYYLNRKKDFNARMGNAVVGQIKLLKQSEEKNLKSNELKNLLDGKKLNVKFDREKPEF
KTKYAKQSYDDFQKRLEERQLAREKNEVHFKKISEKFKDKKLIRKIDRVINDDLNMYRAKNDYEVAQRMI
EQEKMRQAREFEIGMN
MTEHIFNRKSASIILKAKFETGKSKKNNHMVELQKFVDYISRQEAIRQDKLDHDYSEDELNELARIEKAL
EKIEQETDQPVIKDLREMDKYIDYMTRKKAILENVDNEIVNGAFSNTKRYITKKDISKIKESVIEAKNNG
SVMFQDVISFDNDFLVREGYYNPETNELNENVLYEATKSMMGKMQEKEELVDPFWFATIHRNTEHIHIHV
TAMERKNTREIMEYDGVLQARGKRKQSTLDDMIFKFGSKILDRTNEFEKISKLRKEVPLELKQSVKDSLI
QLYVENNSRYDKELVKYLKELKKEIPSTTRGYNELPEETKEKIDEVTNYMTKDNPKRKEYDRMTKEIDEL
YQTTYGKRYNPQEYYLNRKKDFNARMGNAVVGQIKLLKQSEEKNLKSNELKNLLDGKKLNVKFDREKPEF
KTKYAKQSYDDFQKRLEERQLAREKNEVHFKKISEKFKDKKLIRKIDRVINDDLNMYRAKNDYEVAQRMI
EQEKMRQAREFEIGMN
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | B5U8V6 |
T4CP
| ID | 11513 | GenBank | WP_033795274 |
| Name | t4cp2_FKZ20_RS00690_p32 |
UniProt ID | _ |
| Length | 952 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 952 a.a. Molecular weight: 108268.06 Da Isoelectric Point: 8.1346
>WP_033795274.1 VirD4-like conjugal transfer protein, CD1115 family [Enterococcus faecium]
MKLVQNSEVKKRKNGFAKNSNKILVKLKKAGSLKKNKYHTGKIEGEDLPKSLQDKLNVPLSFLGIFLGSI
VFLAITYLLNFLKSLLVAIGQSESGLLGVEFDPKFSLTHLRPLGDTKQYFMCAVVVVIGVAYLIYGKLNH
ESEKNLVYGQKGDSRLSTIKEIKRDYHEIPEKTEVYDGIGGVPISHYQDKYYIDRDTVNTVVLGASRSGK
GETFVVPLLDNLSRAKIQSNMVVNDPKGELFSASKETLERRGYRVLVLNIDDPLESMSFNPLQLVIDSWA
NKDYHEASKRANTLTSMLFASGMGTDNEFFYKSAKSAVNAIILTIVEHCFNNDCIEKITMYNVAQMLNEL
GSLFYTDPKTGKEKNALDEYFNTLPQGNQAKIQYGSTSFAGDKAKGSILSTASQGIEMFTSDLFGKLTSK
MSIDLKEIGFPKSIQFKVNTNLVGKRISISFLRRENDVIRLIKKYRVKVKALGLCVLNFNEYLQDGDMLD
IRYEEEKSRAWYRIEFPKKQANSHDTSLVTKKFKSGANFSDLEISTLRLKYTDQPTAVFMVIPDYDPSNH
VLASIFISQLYTELASNCKQTPDKKCFRRVHFLLDEFGNMPAIDNMDGIMTVCLGRNMLFDLVIQSYSQL
ETRYDKAFKTIKENCQNHILIMSNDEETIEEISKKCGHKTTINRSASSKHLDTDSSVTSSAEQERVITVE
RASQLIEGEQIILRNLHRQDTKRNKIRPFPIFNTKETNMPYRYQFLAEDFDTQRDINEIDIACEHINLSL
QDNQIPYAKFIKDLKTRLEYSIINNIPISDEDYHNYRNLSGSQQTINEEELLKLVNTEEKNQDTEGQVLS
EQEMKINIYLKESQKEAIELVKVINGRLADENSADMVIKTINSQVLDTPFIQSLINDSIAIHNAKENKDQ
IMILEQQLANAYELAVLKTKNSILKNKIIKLKNAMTKLVKAI
MKLVQNSEVKKRKNGFAKNSNKILVKLKKAGSLKKNKYHTGKIEGEDLPKSLQDKLNVPLSFLGIFLGSI
VFLAITYLLNFLKSLLVAIGQSESGLLGVEFDPKFSLTHLRPLGDTKQYFMCAVVVVIGVAYLIYGKLNH
ESEKNLVYGQKGDSRLSTIKEIKRDYHEIPEKTEVYDGIGGVPISHYQDKYYIDRDTVNTVVLGASRSGK
GETFVVPLLDNLSRAKIQSNMVVNDPKGELFSASKETLERRGYRVLVLNIDDPLESMSFNPLQLVIDSWA
NKDYHEASKRANTLTSMLFASGMGTDNEFFYKSAKSAVNAIILTIVEHCFNNDCIEKITMYNVAQMLNEL
GSLFYTDPKTGKEKNALDEYFNTLPQGNQAKIQYGSTSFAGDKAKGSILSTASQGIEMFTSDLFGKLTSK
MSIDLKEIGFPKSIQFKVNTNLVGKRISISFLRRENDVIRLIKKYRVKVKALGLCVLNFNEYLQDGDMLD
IRYEEEKSRAWYRIEFPKKQANSHDTSLVTKKFKSGANFSDLEISTLRLKYTDQPTAVFMVIPDYDPSNH
VLASIFISQLYTELASNCKQTPDKKCFRRVHFLLDEFGNMPAIDNMDGIMTVCLGRNMLFDLVIQSYSQL
ETRYDKAFKTIKENCQNHILIMSNDEETIEEISKKCGHKTTINRSASSKHLDTDSSVTSSAEQERVITVE
RASQLIEGEQIILRNLHRQDTKRNKIRPFPIFNTKETNMPYRYQFLAEDFDTQRDINEIDIACEHINLSL
QDNQIPYAKFIKDLKTRLEYSIINNIPISDEDYHNYRNLSGSQQTINEEELLKLVNTEEKNQDTEGQVLS
EQEMKINIYLKESQKEAIELVKVINGRLADENSADMVIKTINSQVLDTPFIQSLINDSIAIHNAKENKDQ
IMILEQQLANAYELAVLKTKNSILKNKIIKLKNAMTKLVKAI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 15863 | GenBank | NZ_CP041255 |
| Plasmid name | p32 | Incompatibility group | - |
| Plasmid size | 53447 bp | Coordinate of oriT [Strand] | 40063..40255 [+] |
| Host baterium | Enterococcus faecium strain 515 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA1 |