Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 115343 |
| Name | oriT_pNIHE10-0087_oxa |
| Organism | Enterobacter hormaechei strain NIHE10-0087 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_AP025037 (40664..40769 [+], 106 nt) |
| oriT length | 106 nt |
| IRs (inverted repeats) | 30..35, 41..46 (CCGCCG..CGGCGG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 106 nt
>oriT_pNIHE10-0087_oxa
AGTACGGGACAAGATGTGTTTTTGGAGTACCGCCGACACGCGGCGGCCGTCCAAAAACGTCTTGGTCAGGGCAAGCCCCGACACCCCCTAACGAGGTTAGCTATCT
AGTACGGGACAAGATGTGTTTTTGGAGTACCGCCGACACGCGGCGGCCGTCCAAAAACGTCTTGGTCAGGGCAAGCCCCGACACCCCCTAACGAGGTTAGCTATCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file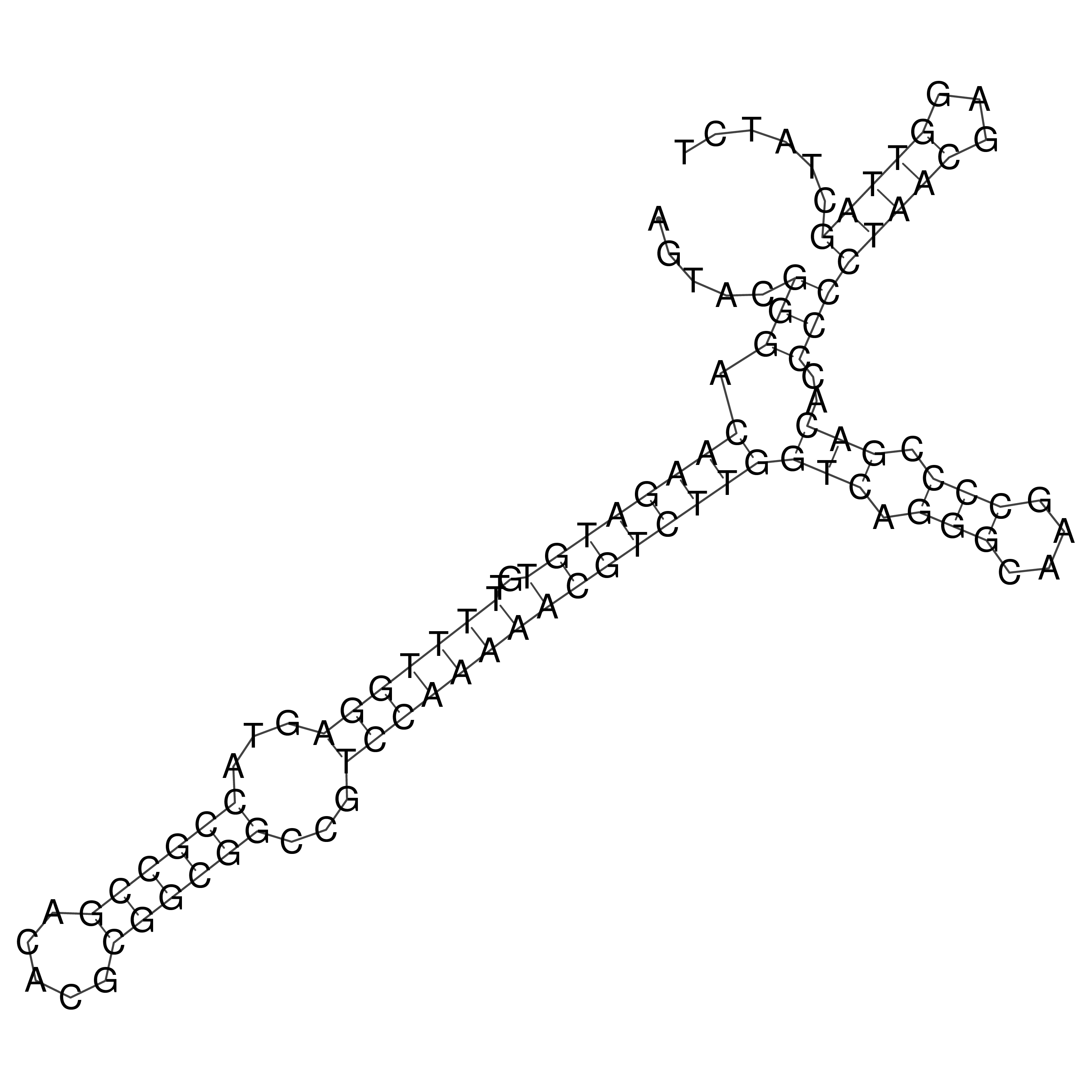
Relaxase
| ID | 9800 | GenBank | WP_004187323 |
| Name | traI_LDO06_RS01195_pNIHE10-0087_oxa |
UniProt ID | A0A3U8KUL3 |
| Length | 659 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 659 a.a. Molecular weight: 75179.89 Da Isoelectric Point: 9.9948
>WP_004187323.1 MULTISPECIES: TraI/MobA(P) family conjugative relaxase [Enterobacterales]
MVIPKIIEGRRDKKSSFGQLIKYMADKPSQELTDTVQPTPETALAVKSDMFEGLNNYLTRKQKVISTPVD
VEPGVQRVVVGDVTCQYNTFSLDGAAQEMNSVSQQSTRCKDPVMHYVLSWPDYEKPNDDQVFDSVKFTLA
SMGMSDHQYVAAIHRDTDNLHVHVAVNRINPQTYKAASSSFTKDTLHQACRLLELKNGWSHSNGAYVVND
RQQIVRNPHSKKERGNWRSLDRINKMENKEGVETLYRYIVGDEQVGGSRQNLIHVSAGLREAKSWDDVHK
TFADIGLRVEKAQGKKGYVITHEHQNQKTAVKASLVFNKAQYTLKSMEERFGEYQPSHIEPAKVSVFKTA
YTPGAYRRDANKRLQRKIERAEERMLLKGRYRAYRNNLPIYSPDKDRIADEYRKIAQHTRLVKNNVRHSV
SDPHTRKLMYNLAEFKRLQAVANLRLSLREERNGFRAANPRLSYREWVEQEALKGDKAALSQMRGFAYSS
RKKEKYKQQLVEQIGFNRTFNAITSHDRDDVAVMASARHGVKPRLLKDGTVIFERDGKPVAADRGHIVLT
ESNGIDKEKTADLAIALTIAGKAKSVRVDGDGEFKELCCNRIVDAAVNHNHPVAQGITFTDAAQQAYAQN
EKHRLIREQNNSKNEMQFRSESDDKFNPK
MVIPKIIEGRRDKKSSFGQLIKYMADKPSQELTDTVQPTPETALAVKSDMFEGLNNYLTRKQKVISTPVD
VEPGVQRVVVGDVTCQYNTFSLDGAAQEMNSVSQQSTRCKDPVMHYVLSWPDYEKPNDDQVFDSVKFTLA
SMGMSDHQYVAAIHRDTDNLHVHVAVNRINPQTYKAASSSFTKDTLHQACRLLELKNGWSHSNGAYVVND
RQQIVRNPHSKKERGNWRSLDRINKMENKEGVETLYRYIVGDEQVGGSRQNLIHVSAGLREAKSWDDVHK
TFADIGLRVEKAQGKKGYVITHEHQNQKTAVKASLVFNKAQYTLKSMEERFGEYQPSHIEPAKVSVFKTA
YTPGAYRRDANKRLQRKIERAEERMLLKGRYRAYRNNLPIYSPDKDRIADEYRKIAQHTRLVKNNVRHSV
SDPHTRKLMYNLAEFKRLQAVANLRLSLREERNGFRAANPRLSYREWVEQEALKGDKAALSQMRGFAYSS
RKKEKYKQQLVEQIGFNRTFNAITSHDRDDVAVMASARHGVKPRLLKDGTVIFERDGKPVAADRGHIVLT
ESNGIDKEKTADLAIALTIAGKAKSVRVDGDGEFKELCCNRIVDAAVNHNHPVAQGITFTDAAQQAYAQN
EKHRLIREQNNSKNEMQFRSESDDKFNPK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3U8KUL3 |
T4CP
| ID | 11425 | GenBank | WP_004206885 |
| Name | t4cp2_LDO06_RS00895_pNIHE10-0087_oxa |
UniProt ID | _ |
| Length | 695 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 695 a.a. Molecular weight: 79538.91 Da Isoelectric Point: 4.8317
>WP_004206885.1 MULTISPECIES: conjugal transfer protein TrbC [Enterobacterales]
MQQESVDSKKVLRPDGSFMEWFLTPNVQFGLLAILTVLGLFLPFTMLLSILILPPLMVAFTDRKFRPPLR
MPKDCEMFDDTLTTETQAEYRLGPIRIPHKVRERKKAKGILYVGYERGRLFGRELWLNMTDLLRHMVFFG
TTGSGKTETFYGFIVNFLLWCRGYCLSDGKADNKLAFATWSLARRFGREDDYYVLNLLTGSIDRFVNLVK
QESIPAQSNSVNLFSVAPPTFIIQLMESMLPQVGGDSAQWQDTAKAMMSALINALCYKRARGELLLSQRT
IQKNMSLPAMAALYVEAKKNGWHSEGYAALESYLENTPGFLLANAEYPETWEARAFEQHNYMSRQFLKTL
SLFNETYGHVFPEDSGDIRMDDIFHNDRILIVMIPSLELSRGEAATLGRLYVTLQRMTISKDLGYQLEGK
KEEVLLTHALNNQAPYGLIYDELGQYFTSGMDTLSAQMRSLEKMGVFSSQDHPSLARGANGEVDSLIANT
RVKYFESIEDRKTFEILRETVGQDYYSELSGQEAEHGTFSKTVREGDLYQIREKDRVNIRELRKQTEGQG
VISFQDALVRSAAFYIPDNEKFSSELPMRINRFIEVLPPSPATLYSIYPERKDDELAETNPDAMRFPPIK
LFGYDKAANSLLEKIEVFYELQCGAISPEEMSVCLFELFAEWLDGTETDDVLYHQEDDLFPMIEM
MQQESVDSKKVLRPDGSFMEWFLTPNVQFGLLAILTVLGLFLPFTMLLSILILPPLMVAFTDRKFRPPLR
MPKDCEMFDDTLTTETQAEYRLGPIRIPHKVRERKKAKGILYVGYERGRLFGRELWLNMTDLLRHMVFFG
TTGSGKTETFYGFIVNFLLWCRGYCLSDGKADNKLAFATWSLARRFGREDDYYVLNLLTGSIDRFVNLVK
QESIPAQSNSVNLFSVAPPTFIIQLMESMLPQVGGDSAQWQDTAKAMMSALINALCYKRARGELLLSQRT
IQKNMSLPAMAALYVEAKKNGWHSEGYAALESYLENTPGFLLANAEYPETWEARAFEQHNYMSRQFLKTL
SLFNETYGHVFPEDSGDIRMDDIFHNDRILIVMIPSLELSRGEAATLGRLYVTLQRMTISKDLGYQLEGK
KEEVLLTHALNNQAPYGLIYDELGQYFTSGMDTLSAQMRSLEKMGVFSSQDHPSLARGANGEVDSLIANT
RVKYFESIEDRKTFEILRETVGQDYYSELSGQEAEHGTFSKTVREGDLYQIREKDRVNIRELRKQTEGQG
VISFQDALVRSAAFYIPDNEKFSSELPMRINRFIEVLPPSPATLYSIYPERKDDELAETNPDAMRFPPIK
LFGYDKAANSLLEKIEVFYELQCGAISPEEMSVCLFELFAEWLDGTETDDVLYHQEDDLFPMIEM
Protein domains
No domain identified.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 43565..62764
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LDO06_RS01175 (NIHE100087_pO0600) | 38649..38960 | + | 312 | WP_004187333 | hypothetical protein | - |
| LDO06_RS01180 (NIHE100087_pO0610) | 39093..39629 | + | 537 | WP_004187332 | hypothetical protein | - |
| LDO06_RS01185 (NIHE100087_pO0620) | 40131..40526 | - | 396 | WP_179213477 | MobC | - |
| LDO06_RS01190 (NIHE100087_pO0630) | 40561..41088 | + | 528 | WP_172740549 | plasmid mobilization protein MobA | - |
| LDO06_RS01195 (NIHE100087_pO0640) | 41075..43054 | + | 1980 | WP_004187323 | TraI/MobA(P) family conjugative relaxase | - |
| LDO06_RS01200 (NIHE100087_pO0650) | 43068..43568 | + | 501 | WP_004187320 | DotD/TraH family lipoprotein | - |
| LDO06_RS01205 (NIHE100087_pO0660) | 43565..44344 | + | 780 | WP_004187315 | type IV secretory system conjugative DNA transfer family protein | traI |
| LDO06_RS01210 (NIHE100087_pO0670) | 44355..45518 | + | 1164 | WP_004187313 | plasmid transfer ATPase TraJ | virB11 |
| LDO06_RS01215 (NIHE100087_pO0680) | 45508..45768 | + | 261 | WP_004187310 | IcmT/TraK family protein | traK |
| LDO06_RS01220 (NIHE100087_pO0690) | 45793..49140 | + | 3348 | WP_223963185 | LPD7 domain-containing protein | - |
| LDO06_RS01225 (NIHE100087_pO0700) | 49106..49618 | + | 513 | WP_011091071 | hypothetical protein | traL |
| LDO06_RS01230 (NIHE100087_pO0710) | 49619..49831 | - | 213 | WP_305953721 | Hha/YmoA family nucleoid-associated regulatory protein | - |
| LDO06_RS01235 (NIHE100087_pO0720) | 49803..50213 | - | 411 | WP_004187465 | H-NS family nucleoid-associated regulatory protein | - |
| LDO06_RS01240 (NIHE100087_pO0730) | 50281..50994 | + | 714 | WP_004187467 | DotI/IcmL family type IV secretion protein | traM |
| LDO06_RS01245 (NIHE100087_pO0740) | 51003..52154 | + | 1152 | WP_004187471 | DotH/IcmK family type IV secretion protein | traN |
| LDO06_RS01250 (NIHE100087_pO0750) | 52166..53515 | + | 1350 | WP_004187474 | conjugal transfer protein TraO | traO |
| LDO06_RS01255 (NIHE100087_pO0760) | 53527..54231 | + | 705 | WP_015060002 | conjugal transfer protein TraP | traP |
| LDO06_RS01260 (NIHE100087_pO0770) | 54255..54785 | + | 531 | WP_004187478 | conjugal transfer protein TraQ | traQ |
| LDO06_RS01265 (NIHE100087_pO0780) | 54802..55191 | + | 390 | WP_004187479 | DUF6750 family protein | traR |
| LDO06_RS01270 (NIHE100087_pO0790) | 55237..55731 | + | 495 | WP_004187480 | hypothetical protein | - |
| LDO06_RS01275 (NIHE100087_pO0800) | 55728..58778 | + | 3051 | WP_004187482 | hypothetical protein | traU |
| LDO06_RS01280 (NIHE100087_pO0810) | 58775..59983 | + | 1209 | WP_011091082 | conjugal transfer protein TraW | traW |
| LDO06_RS01285 (NIHE100087_pO0820) | 59980..60576 | + | 597 | WP_015060003 | hypothetical protein | - |
| LDO06_RS01290 (NIHE100087_pO0830) | 60569..62764 | + | 2196 | WP_015062834 | DotA/TraY family protein | traY |
| LDO06_RS01295 (NIHE100087_pO0840) | 62766..63419 | + | 654 | WP_015060005 | hypothetical protein | - |
Host bacterium
| ID | 15777 | GenBank | NZ_AP025037 |
| Plasmid name | pNIHE10-0087_oxa | Incompatibility group | IncL/M |
| Plasmid size | 63499 bp | Coordinate of oriT [Strand] | 40664..40769 [+] |
| Host baterium | Enterobacter hormaechei strain NIHE10-0087 |
Cargo genes
| Drug resistance gene | blaOXA-48 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |