Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 115220 |
| Name | oriT_pC-CRECL423 |
| Organism | Enterobacter hormaechei strain CRECL423 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP133339 (48512..48662 [+], 151 nt) |
| oriT length | 151 nt |
| IRs (inverted repeats) | 93..98, 106..111 (TGGCCT..AGGCCA) 12..17, 24..29 (AACCCT..AGGGTT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 151 nt
>oriT_pC-CRECL423
TGGTCGATGTGAACCCTTTCGACAGGGTTATGAATGAATTGAAAAGTCGTGGCCGCAAGAACGCTCACATCCTGAGCATCCTCCAATTCGACTGGCCTGCATCGGAGGCCATCATCGAGAAGCTGAGCTGCTACATCACAGACGGGATTAA
TGGTCGATGTGAACCCTTTCGACAGGGTTATGAATGAATTGAAAAGTCGTGGCCGCAAGAACGCTCACATCCTGAGCATCCTCCAATTCGACTGGCCTGCATCGGAGGCCATCATCGAGAAGCTGAGCTGCTACATCACAGACGGGATTAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file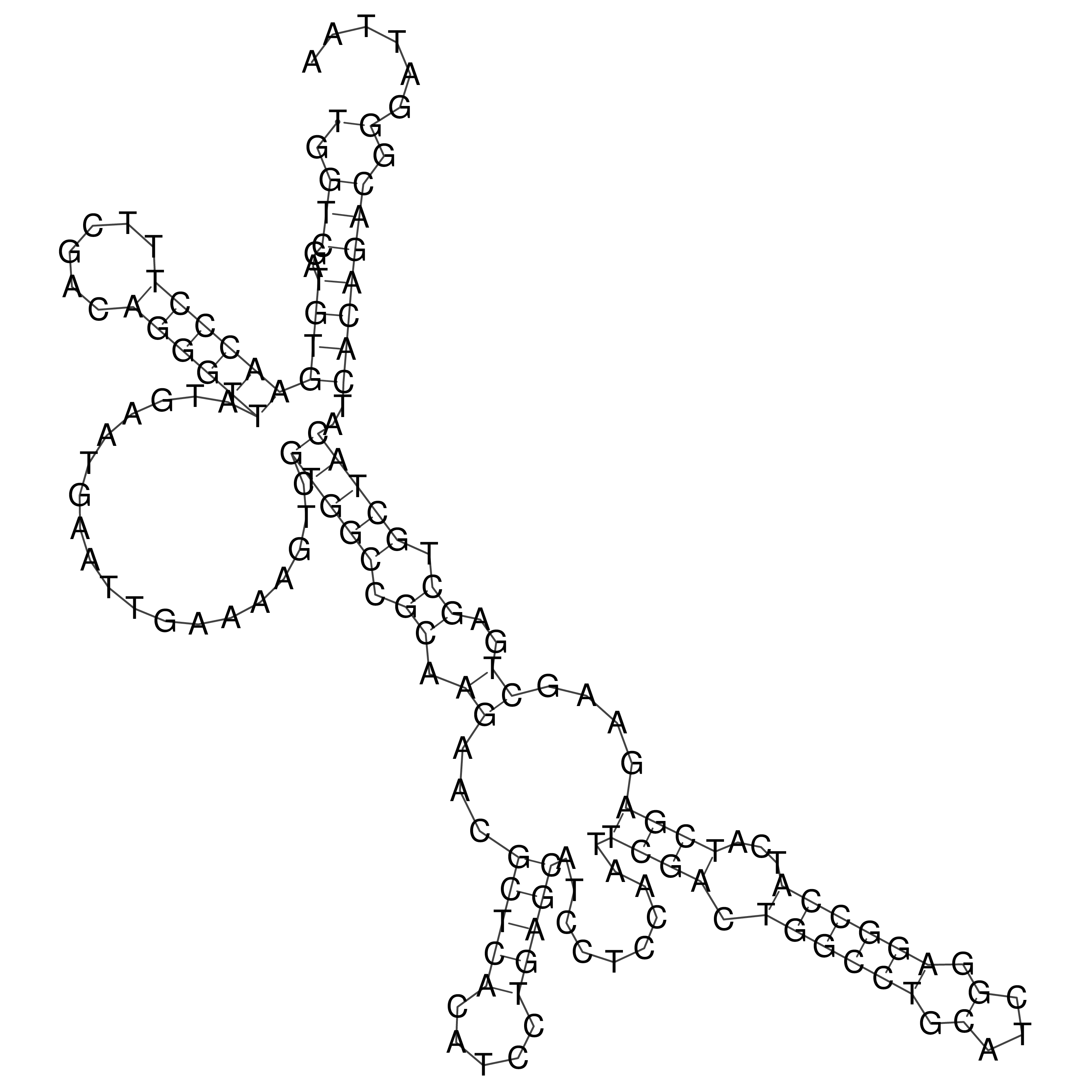
Relaxase
| ID | 9726 | GenBank | WP_308982204 |
| Name | mobH_RCR42_RS25820_pC-CRECL423 |
UniProt ID | _ |
| Length | 990 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 990 a.a. Molecular weight: 109482.12 Da Isoelectric Point: 4.9964
>WP_308982204.1 MobH family relaxase [Enterobacter hormaechei]
MLKALNKLFGGRSGVIETAPSVRVLPLKDVEDEEIPRYPPFAKGLPVAPLDKILATQAELIEKVRNSLGF
TVDDFNRLVLPVIQRYAAFVHLLPASESHHHRGAGGLFRHGLEVAFWAAQASESVIFSIEGTPRERRDNE
PRWRLASCFSGLLHDVGKPLSDVSITDKDGSITWNPYSESLHDWAHRHEIDRYFIRWRDKRHKRHEQFSL
LAVDRIIPAETREFLSKSGPSIMEAMLEAISGTSVNQPVTKLMLRADQESVSRDLRQSRLDVDEFSYGVP
VERYVFDAIRRLVKTGKWKVNEPGAKVWHLNQGVFIAWKQLGDLYDLISHDKIPGIPRDPDTLADILIER
GFAVPNTVQEKGERAYYRYWEVLPEMLQEAAGSVKILMLRLESNDLVFTTEPPAAVAAEVVGDVEDAEIE
FVDPEEVDDDQEEDVSALNDDMLAAEQEAEKALAGLGFGDAMEMLKSTSDAVEEKPEQKDAGSTESSKPD
AGKKGKPQSKPGKAKPKSDTEKQPHKPEAKEDLSPQDIAKNAPPLANDNPLQALKDVGGGLGDIDFPFDA
FSASAETTSTDATNSEIPDVAMPGKQEKQPKQDFVPQEQNSLQGDDFPMFGSSDEPPSWAIEPLPMLTDA
PEQTTPAPAMPPTDKPNLHEKDAKTLLVEMLAGYGEASALLEQAIMPVLEGKTTLGEVLCLMKGQAVILY
PDGARSLGAPSEVLSKLSHANAIVPDPIMPGRKVRDFSGVKAIVLAEQLSDAVVAAIKDAEASMGGYQDA
FELVSPPRLDANKNKSAPKQQSRKKAQQQKPEVNAGKPSPEQKAKGKDSQPQPKEKKVDVTSPVEEPQRK
PVQEKQNVARLPKREVQPVAPEPKVEREKELGHVEVREREEPEVREFEPPKAKTNPKDINAEDFLPSGVT
PQKALQMLKDMIQKRSGRWLVTPVLEEDGCLVTSDKAFDMIAGENIGISKHILCGMLSRAQRRPLLKKRQ
GKLYLEVNET
MLKALNKLFGGRSGVIETAPSVRVLPLKDVEDEEIPRYPPFAKGLPVAPLDKILATQAELIEKVRNSLGF
TVDDFNRLVLPVIQRYAAFVHLLPASESHHHRGAGGLFRHGLEVAFWAAQASESVIFSIEGTPRERRDNE
PRWRLASCFSGLLHDVGKPLSDVSITDKDGSITWNPYSESLHDWAHRHEIDRYFIRWRDKRHKRHEQFSL
LAVDRIIPAETREFLSKSGPSIMEAMLEAISGTSVNQPVTKLMLRADQESVSRDLRQSRLDVDEFSYGVP
VERYVFDAIRRLVKTGKWKVNEPGAKVWHLNQGVFIAWKQLGDLYDLISHDKIPGIPRDPDTLADILIER
GFAVPNTVQEKGERAYYRYWEVLPEMLQEAAGSVKILMLRLESNDLVFTTEPPAAVAAEVVGDVEDAEIE
FVDPEEVDDDQEEDVSALNDDMLAAEQEAEKALAGLGFGDAMEMLKSTSDAVEEKPEQKDAGSTESSKPD
AGKKGKPQSKPGKAKPKSDTEKQPHKPEAKEDLSPQDIAKNAPPLANDNPLQALKDVGGGLGDIDFPFDA
FSASAETTSTDATNSEIPDVAMPGKQEKQPKQDFVPQEQNSLQGDDFPMFGSSDEPPSWAIEPLPMLTDA
PEQTTPAPAMPPTDKPNLHEKDAKTLLVEMLAGYGEASALLEQAIMPVLEGKTTLGEVLCLMKGQAVILY
PDGARSLGAPSEVLSKLSHANAIVPDPIMPGRKVRDFSGVKAIVLAEQLSDAVVAAIKDAEASMGGYQDA
FELVSPPRLDANKNKSAPKQQSRKKAQQQKPEVNAGKPSPEQKAKGKDSQPQPKEKKVDVTSPVEEPQRK
PVQEKQNVARLPKREVQPVAPEPKVEREKELGHVEVREREEPEVREFEPPKAKTNPKDINAEDFLPSGVT
PQKALQMLKDMIQKRSGRWLVTPVLEEDGCLVTSDKAFDMIAGENIGISKHILCGMLSRAQRRPLLKKRQ
GKLYLEVNET
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 11343 | GenBank | WP_000178857 |
| Name | traD_RCR42_RS25825_pC-CRECL423 |
UniProt ID | _ |
| Length | 621 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 621 a.a. Molecular weight: 69225.71 Da Isoelectric Point: 6.9403
>WP_000178857.1 MULTISPECIES: conjugative transfer system coupling protein TraD [Gammaproteobacteria]
MTMSYDPLAYEMPWRPNYEKNAVAGWLAASGAALAVEQVSTMPPEPFYWMTGICGVMAMARLPKAIKLHL
LQKHLKGRDLEFISIAELQKYIKDTPDDMWLGSGFLWENRHAQRVFEILKRDWTSIVGRESTVKKVVRKI
QGKKKELPIGQPWIHGVEPKEEKLMQPLKHTEGHSLIVGTTGSGKTRMFDILISQAILRGEAVIIIDPKG
DKEMRDNARRACEAMGQPERFVSFHPAFPEESVRIDPLRNFTRVTEIASRLAALIPSEAGADPFKSFGWQ
ALNNIAQGLVITHDRPNLTKLRRFLEGGAAGLVIKAVQAYSERVMPDWEAEAAAYLEKVKNGSREKIAFA
LMKFYYDIIQPEHPNSDLEGLLSMFQHDQTHFSKMVANLLPIMNMLTSGELGPLLSPDSSDLSDERQITD
SAKIINNAQVAYLGLDSLTDNMVGSAMGSIFLSDLTAVAGDRYNYGVNNRPVNIFVDEAAEVINDPFIQL
LNKGRGAKLRLFVATQTFADFAARLGSKDKALQVLGNINNTFALRIVDGETQEYIADNLPKTRLKYVMRT
QGQNSDGKEPIMHGGNQGERLMEEEADLFPAQLLGMLPNLEYIAKISGGTIVKGRLPILTQ
MTMSYDPLAYEMPWRPNYEKNAVAGWLAASGAALAVEQVSTMPPEPFYWMTGICGVMAMARLPKAIKLHL
LQKHLKGRDLEFISIAELQKYIKDTPDDMWLGSGFLWENRHAQRVFEILKRDWTSIVGRESTVKKVVRKI
QGKKKELPIGQPWIHGVEPKEEKLMQPLKHTEGHSLIVGTTGSGKTRMFDILISQAILRGEAVIIIDPKG
DKEMRDNARRACEAMGQPERFVSFHPAFPEESVRIDPLRNFTRVTEIASRLAALIPSEAGADPFKSFGWQ
ALNNIAQGLVITHDRPNLTKLRRFLEGGAAGLVIKAVQAYSERVMPDWEAEAAAYLEKVKNGSREKIAFA
LMKFYYDIIQPEHPNSDLEGLLSMFQHDQTHFSKMVANLLPIMNMLTSGELGPLLSPDSSDLSDERQITD
SAKIINNAQVAYLGLDSLTDNMVGSAMGSIFLSDLTAVAGDRYNYGVNNRPVNIFVDEAAEVINDPFIQL
LNKGRGAKLRLFVATQTFADFAARLGSKDKALQVLGNINNTFALRIVDGETQEYIADNLPKTRLKYVMRT
QGQNSDGKEPIMHGGNQGERLMEEEADLFPAQLLGMLPNLEYIAKISGGTIVKGRLPILTQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 11344 | GenBank | WP_088212670 |
| Name | traC_RCR42_RS25900_pC-CRECL423 |
UniProt ID | _ |
| Length | 815 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 815 a.a. Molecular weight: 92632.91 Da Isoelectric Point: 5.9216
>WP_088212670.1 MULTISPECIES: type IV secretion system protein TraC [Gammaproteobacteria]
MIVTIKKKLEETLIPEHLRAAGIIPVLAYDEDDHVFLMDDHSAGFGFMCEPLCGADEKVQERMNGFLNQE
FPSKTTLQFVLFRSPDINQEMYRMMGLRDGFRHELLTSVIKERINFLQHHTTDRIFAKTNKGIYDNGLIQ
DLKLFVTCKVPIKNNNPTESELQQLAQLRTKVESSLQTVGLRPRTMTAVNYIRIMSTILNWGPDASWRHD
SVDWEMDKPICEQIFDYGTDVEVSKNGIRLGDYHAKVMSAKKLPDVFYFGDALTYVGDLSGGNSSIKENY
MVVTNVFFPEAESTKNTLERKRQFTVNQAYGPMLKFVPVLADKKESFDTLYESMKEGAKPVKITYSVVLF
APTKERVEAAAMAARNIWRESRFELMEDKFVALPMFLNCLPFCTDRDAVRDLFRYKTMTTEQAAVVLPVF
GEWKGTGTYHAALISRNGQLMSLSLHDSNTNKNLVIAAESGSGKSFLTNELIFSYLSEGAQVWVIDAGKS
YQKLSEMLNGDFVHFEEGTHVCLNPFELIQNYEDEEDAIVSLVCAMASAKGLLDEWQISALKQVLSRLWE
EKGKEMKVDDIAERCLEEENDQRLKDIGQQLYAFTSQGSYGKYFSRKNNVSFQNQFTVLELDELQGRKHL
RQVVLLQLIYQIQQEVFLGERNRKKVVIVDEAWDLLKEGEVSVFMEHAYRKFRKYGGSVVIATQSINDLY
ENAVGRAIAENSASMYLLGQTEETVESVKRSGRLTLSEGGFHTLKTVHTIQGVYSEIFIKSKSGMGVGRL
IVGDFQKLLYSTDPVDVNAIDQFVKQGMSIPEAIKAVMRSRRQAA
MIVTIKKKLEETLIPEHLRAAGIIPVLAYDEDDHVFLMDDHSAGFGFMCEPLCGADEKVQERMNGFLNQE
FPSKTTLQFVLFRSPDINQEMYRMMGLRDGFRHELLTSVIKERINFLQHHTTDRIFAKTNKGIYDNGLIQ
DLKLFVTCKVPIKNNNPTESELQQLAQLRTKVESSLQTVGLRPRTMTAVNYIRIMSTILNWGPDASWRHD
SVDWEMDKPICEQIFDYGTDVEVSKNGIRLGDYHAKVMSAKKLPDVFYFGDALTYVGDLSGGNSSIKENY
MVVTNVFFPEAESTKNTLERKRQFTVNQAYGPMLKFVPVLADKKESFDTLYESMKEGAKPVKITYSVVLF
APTKERVEAAAMAARNIWRESRFELMEDKFVALPMFLNCLPFCTDRDAVRDLFRYKTMTTEQAAVVLPVF
GEWKGTGTYHAALISRNGQLMSLSLHDSNTNKNLVIAAESGSGKSFLTNELIFSYLSEGAQVWVIDAGKS
YQKLSEMLNGDFVHFEEGTHVCLNPFELIQNYEDEEDAIVSLVCAMASAKGLLDEWQISALKQVLSRLWE
EKGKEMKVDDIAERCLEEENDQRLKDIGQQLYAFTSQGSYGKYFSRKNNVSFQNQFTVLELDELQGRKHL
RQVVLLQLIYQIQQEVFLGERNRKKVVIVDEAWDLLKEGEVSVFMEHAYRKFRKYGGSVVIATQSINDLY
ENAVGRAIAENSASMYLLGQTEETVESVKRSGRLTLSEGGFHTLKTVHTIQGVYSEIFIKSKSGMGVGRL
IVGDFQKLLYSTDPVDVNAIDQFVKQGMSIPEAIKAVMRSRRQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 46596..71742
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| RCR42_RS25810 (RCR42_25810) | 42627..42860 | - | 234 | WP_001191890 | hypothetical protein | - |
| RCR42_RS25815 (RCR42_25815) | 42842..43459 | - | 618 | WP_001249395 | hypothetical protein | - |
| RCR42_RS25820 (RCR42_25820) | 43627..46599 | + | 2973 | WP_308982204 | MobH family relaxase | - |
| RCR42_RS25825 (RCR42_25825) | 46596..48461 | + | 1866 | WP_000178857 | conjugative transfer system coupling protein TraD | virb4 |
| RCR42_RS25830 (RCR42_25830) | 48511..49056 | + | 546 | WP_247750980 | hypothetical protein | - |
| RCR42_RS25835 (RCR42_25835) | 49013..49642 | + | 630 | WP_088212662 | DUF4400 domain-containing protein | tfc7 |
| RCR42_RS25840 (RCR42_25840) | 49652..50098 | + | 447 | WP_000122507 | hypothetical protein | - |
| RCR42_RS25845 (RCR42_25845) | 50108..50485 | + | 378 | WP_088212663 | hypothetical protein | - |
| RCR42_RS25850 (RCR42_25850) | 50485..51147 | + | 663 | WP_308982208 | hypothetical protein | - |
| RCR42_RS25855 (RCR42_25855) | 51483..51848 | + | 366 | WP_001052531 | hypothetical protein | - |
| RCR42_RS25860 (RCR42_25860) | 51993..52274 | + | 282 | WP_088212665 | type IV conjugative transfer system protein TraL | traL |
| RCR42_RS25865 (RCR42_25865) | 52271..52897 | + | 627 | WP_001049717 | TraE/TraK family type IV conjugative transfer system protein | traE |
| RCR42_RS25870 (RCR42_25870) | 52881..53798 | + | 918 | WP_088212666 | type-F conjugative transfer system secretin TraK | traK |
| RCR42_RS25875 (RCR42_25875) | 53798..55111 | + | 1314 | WP_308982260 | TrbI/VirB10 family protein | traB |
| RCR42_RS25880 (RCR42_25880) | 55108..55686 | + | 579 | WP_088212668 | type IV conjugative transfer system lipoprotein TraV | traV |
| RCR42_RS25885 (RCR42_25885) | 55690..56082 | + | 393 | WP_000479535 | TraA family conjugative transfer protein | - |
| RCR42_RS25890 (RCR42_25890) | 56283..61769 | + | 5487 | WP_308982212 | Ig-like domain-containing protein | - |
| RCR42_RS25895 (RCR42_25895) | 61918..62625 | + | 708 | WP_001259346 | DsbC family protein | - |
| RCR42_RS25900 (RCR42_25900) | 62622..65069 | + | 2448 | WP_088212670 | type IV secretion system protein TraC | virb4 |
| RCR42_RS25905 (RCR42_25905) | 65084..65401 | + | 318 | WP_000351984 | hypothetical protein | - |
| RCR42_RS25910 (RCR42_25910) | 65398..65928 | + | 531 | WP_001010738 | S26 family signal peptidase | - |
| RCR42_RS25915 (RCR42_25915) | 65891..67156 | + | 1266 | WP_088212671 | TrbC family F-type conjugative pilus assembly protein | traW |
| RCR42_RS25920 (RCR42_25920) | 67153..67824 | + | 672 | WP_088212672 | EAL domain-containing protein | - |
| RCR42_RS25925 (RCR42_25925) | 67824..68828 | + | 1005 | WP_005507569 | TraU family protein | traU |
| RCR42_RS25930 (RCR42_25930) | 68944..71742 | + | 2799 | WP_204260578 | conjugal transfer mating pair stabilization protein TraN | traN |
| RCR42_RS25935 (RCR42_25935) | 71782..72642 | - | 861 | WP_088212674 | hypothetical protein | - |
| RCR42_RS25940 (RCR42_25940) | 72765..73406 | - | 642 | WP_088212675 | hypothetical protein | - |
| RCR42_RS25945 (RCR42_25945) | 73702..74022 | + | 321 | WP_163440224 | hypothetical protein | - |
| RCR42_RS25950 (RCR42_25950) | 74326..74511 | + | 186 | WP_001186916 | hypothetical protein | - |
| RCR42_RS25955 (RCR42_25955) | 74731..75699 | + | 969 | WP_113620223 | AAA family ATPase | - |
| RCR42_RS25960 (RCR42_25960) | 75710..76615 | + | 906 | WP_308982217 | hypothetical protein | - |
Host bacterium
| ID | 15655 | GenBank | NZ_CP133339 |
| Plasmid name | pC-CRECL423 | Incompatibility group | IncA/C2 |
| Plasmid size | 147899 bp | Coordinate of oriT [Strand] | 48512..48662 [+] |
| Host baterium | Enterobacter hormaechei strain CRECL423 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |