Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 115219 |
| Name | oriT_pB-CRECL423 |
| Organism | Enterobacter hormaechei strain CRECL423 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP133338 (83324..83426 [+], 103 nt) |
| oriT length | 103 nt |
| IRs (inverted repeats) | 89..94, 96..101 (AAATAA..TTATTT) 28..33, 40..45 (GCAAAA..TTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 103 nt
>oriT_pB-CRECL423
CTCACCACCAAAAGTTCCACACCCCACGCAAAATTAAAGTTTTGCTGATTGGGTATTCAAATCATGCAGTTAGTGAAAAAGTAATGTGAAATAATTTATTTTA
CTCACCACCAAAAGTTCCACACCCCACGCAAAATTAAAGTTTTGCTGATTGGGTATTCAAATCATGCAGTTAGTGAAAAAGTAATGTGAAATAATTTATTTTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file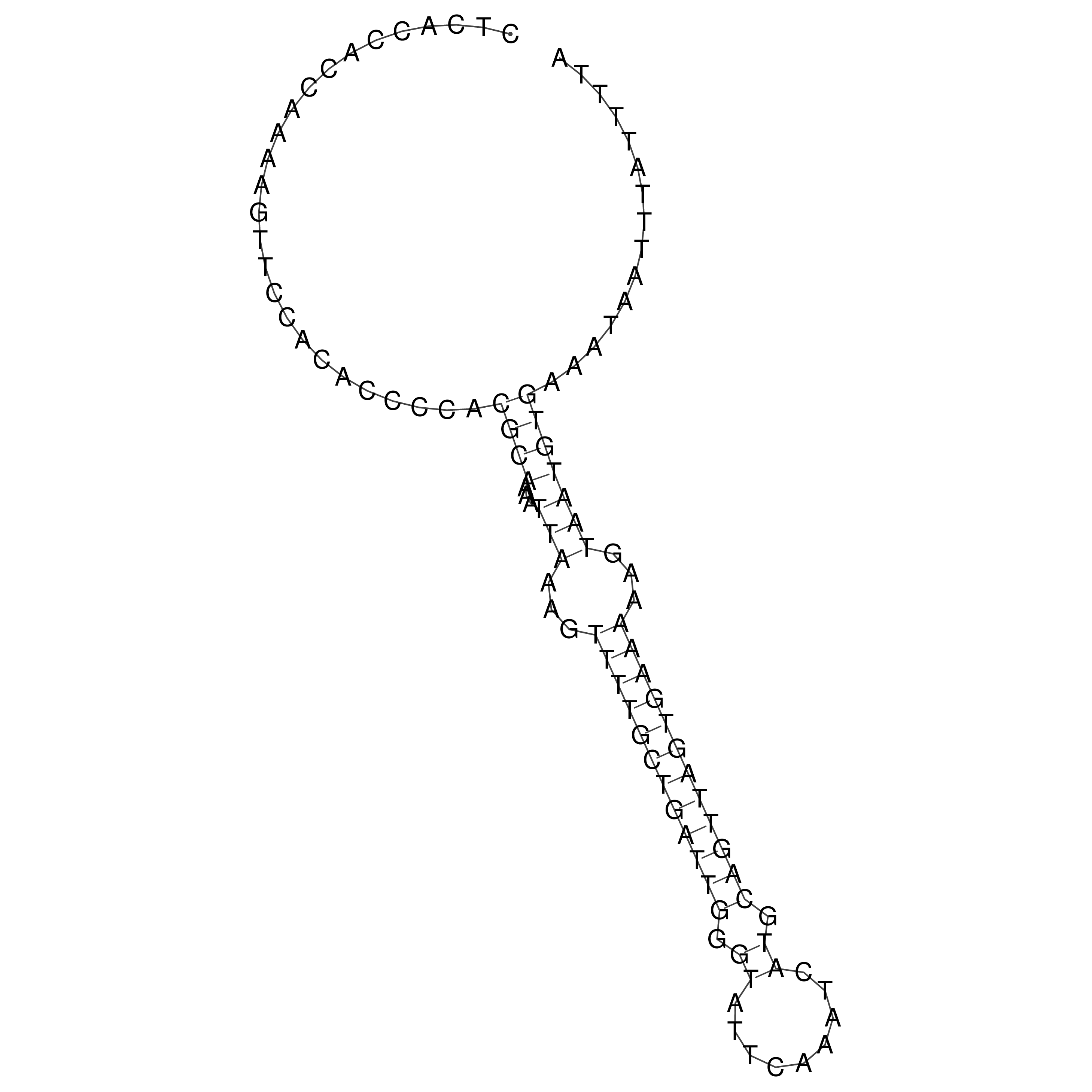
Relaxase
| ID | 9725 | GenBank | WP_016479978 |
| Name | traI_RCR42_RS25240_pB-CRECL423 |
UniProt ID | A0A0G3B767 |
| Length | 1746 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1746 a.a. Molecular weight: 189285.41 Da Isoelectric Point: 6.0099
>WP_016479978.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacterales]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGKGAEALGLSGGVDQKVFTKVLEGRLPDGSDLSRTQ
DGANKHRPGYDFTFSAPKSVSVMAMLGDDKRLIEAHNRAVETAIKQIEAMASTRVMTEGRSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANATQHGDEWRTLSSDKVGKTGFIENIYANQIAFGRLYRAALKDDVAA
MGYETETVGKHGMWELKGVPTEPYSSRSRTISEAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMQT
LKETGFDIKAYREAADLRVVQGNIPATTPEAIDINSSVGQAIAMLSDRRARFTYSELLATTLGQLPARSG
MVEMARDGIDAAIKNEQLIPLDKEKGLFTSNIHVLDELSVSAMTRDLQRTGQADIFPDKSVPRSRSYSDA
VSVLAQDRPPVAIISGQGGAAGQRERVAELTMMAREQGRDVQIIAADRRSRSALMQDERLSGEHITDRRG
LTEGMAFTPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTQVSVISEPDRRQRYDRLAADFARSVRAGEHSVAQVTGPREQAVLAGVVREALKAEKVLGEREVSIT
TLEPVWLDSRHQQVRDHYREGMVMERWNAEERSRERFVIDRVTGRNNSLTLRNAQGETRVTRLSELNSSW
SLYRTGTLQVAEGDRLAVLGQAQGTRLRGGDTVTVKALGEAGLVVSLPGRKADVVLPAGDSPFTAAKVVQ
GWVESPGRSVSDSATVFASLSQRELDNTTLNKLALSGRAVRLYSAQTAQKTTEKLSRQSAWSVVSEQIKD
AAGHDRLDDALAHQKAGLHTPEQQAIHLAIPVLEGNGLAFTKPQLMAAAKEFEQDSLSLQAIEKEADRQV
RSGDLLSVPVSPGNGLQLLVSRQSYNAEKSIIRHVLEGKEAVTPLMDKVPDAQLAGLTDGQRNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEREQPRVIGVAPTHRAVSEMRDAGVPETQTLAAFIHDTQ
QQLRGGERTDFSNVLFLVDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DVAVMQEIVRQTPELKPAVYSLISRDIGSALTTIESVAPAQVPRRADAWQPDSSVMEFSREREEAIAKAV
AAGDLMPGGQPATLLEAIVKDYTGRTSEAQAQTIVITALNADRRQVNAMIHDARQGAGEVGEKEVTLPVL
TPANIRDGELRRMATWEASRDSLVLLDSTYYSIEALDGEAHLVMLKDAQGNTRSLSPAQAATEGVTLYRQ
DTITVSEGDRMRFSKSDNERGFVANSVWSVSEIKGDSVTLSDGKQTRTLTPSAEQAEQHIDLAYAVTVEG
SQGASEPFSISLQGTDGGRKRMVSFESAYVALSRMKQHAQVYTDNRDKWAAAMEKSQAKSTAHDILEPRG
DRAVANAARLTATAKALGEVPAGRAALRQAGLQPEGSMAKYISPGRKYPQPHVALPAFDRNGRKAGVWLS
ALTSGDGQLKGLAGEGRVMGSGDAAFAGLQASRNGESLLARDMEEGVRLARENPQSGVVVRLEGEARPWN
PGAITGGRVWADGLPDATGTQPAETVPPEVLAQQAREAEVQRELDKRAEEAVREMARSGDRPAGGADVAV
RDVVRDMDRIKETPASPLTLPDAPEERRRDEAVSQVVRESVQRDRLQQMERETVRDLEREKTLGGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGKGAEALGLSGGVDQKVFTKVLEGRLPDGSDLSRTQ
DGANKHRPGYDFTFSAPKSVSVMAMLGDDKRLIEAHNRAVETAIKQIEAMASTRVMTEGRSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANATQHGDEWRTLSSDKVGKTGFIENIYANQIAFGRLYRAALKDDVAA
MGYETETVGKHGMWELKGVPTEPYSSRSRTISEAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMQT
LKETGFDIKAYREAADLRVVQGNIPATTPEAIDINSSVGQAIAMLSDRRARFTYSELLATTLGQLPARSG
MVEMARDGIDAAIKNEQLIPLDKEKGLFTSNIHVLDELSVSAMTRDLQRTGQADIFPDKSVPRSRSYSDA
VSVLAQDRPPVAIISGQGGAAGQRERVAELTMMAREQGRDVQIIAADRRSRSALMQDERLSGEHITDRRG
LTEGMAFTPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTQVSVISEPDRRQRYDRLAADFARSVRAGEHSVAQVTGPREQAVLAGVVREALKAEKVLGEREVSIT
TLEPVWLDSRHQQVRDHYREGMVMERWNAEERSRERFVIDRVTGRNNSLTLRNAQGETRVTRLSELNSSW
SLYRTGTLQVAEGDRLAVLGQAQGTRLRGGDTVTVKALGEAGLVVSLPGRKADVVLPAGDSPFTAAKVVQ
GWVESPGRSVSDSATVFASLSQRELDNTTLNKLALSGRAVRLYSAQTAQKTTEKLSRQSAWSVVSEQIKD
AAGHDRLDDALAHQKAGLHTPEQQAIHLAIPVLEGNGLAFTKPQLMAAAKEFEQDSLSLQAIEKEADRQV
RSGDLLSVPVSPGNGLQLLVSRQSYNAEKSIIRHVLEGKEAVTPLMDKVPDAQLAGLTDGQRNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEREQPRVIGVAPTHRAVSEMRDAGVPETQTLAAFIHDTQ
QQLRGGERTDFSNVLFLVDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DVAVMQEIVRQTPELKPAVYSLISRDIGSALTTIESVAPAQVPRRADAWQPDSSVMEFSREREEAIAKAV
AAGDLMPGGQPATLLEAIVKDYTGRTSEAQAQTIVITALNADRRQVNAMIHDARQGAGEVGEKEVTLPVL
TPANIRDGELRRMATWEASRDSLVLLDSTYYSIEALDGEAHLVMLKDAQGNTRSLSPAQAATEGVTLYRQ
DTITVSEGDRMRFSKSDNERGFVANSVWSVSEIKGDSVTLSDGKQTRTLTPSAEQAEQHIDLAYAVTVEG
SQGASEPFSISLQGTDGGRKRMVSFESAYVALSRMKQHAQVYTDNRDKWAAAMEKSQAKSTAHDILEPRG
DRAVANAARLTATAKALGEVPAGRAALRQAGLQPEGSMAKYISPGRKYPQPHVALPAFDRNGRKAGVWLS
ALTSGDGQLKGLAGEGRVMGSGDAAFAGLQASRNGESLLARDMEEGVRLARENPQSGVVVRLEGEARPWN
PGAITGGRVWADGLPDATGTQPAETVPPEVLAQQAREAEVQRELDKRAEEAVREMARSGDRPAGGADVAV
RDVVRDMDRIKETPASPLTLPDAPEERRRDEAVSQVVRESVQRDRLQQMERETVRDLEREKTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 11341 | GenBank | WP_008324134 |
| Name | traC_RCR42_RS25155_pB-CRECL423 |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 98841.51 Da Isoelectric Point: 5.7676
>WP_008324134.1 MULTISPECIES: type IV secretion system protein TraC [Enterobacterales]
MSNPLDSVTRTVNSLLTALKMPDASSEANDVLGQMRFPQFSRVLPYRDYDAETGLFMNEKTMGFMLEAQP
INGANETIVSSLENLMRTKLPREVPVAVHLVSSHLVGQDIEYGLRELSWTGEKADTFNAITRAYYLRAAE
EGFPLPDHLNLPLTLRNYRVYISCCVPRKRGSKASVMEMENQVKIIRASLQGAQIATRSVDADAFIRIAG
EMINHNPDALYPTERELDPLKDLNYQCVDDTFDLKVKADYMTLGLRGNGRNSTARIMNFQLTKNPELAFL
WNMADNYSNLLNPELSVSCPFILTLVLSVEDQVKAQNEANLKFMDLEKKSKTSYAKFFPGVAQEAKEWGD
LRQRLSSGQSSLVSYFLNVTLFCRDDQETALKVEQDVLNSFRKNGFELISPRFNHMRNFLVNLPFMAGGG
LFKQLRESGVLQRAESFNVANLMPVVADSPLATSGLLAPTYRNQLAFIDLFSRGMGNTNFNMAVCGTSGA
GKTGLIQPLIRSVIDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLKFNPFANVTDDTIDEMAERLRDQL
SVMASPNGNLDEVHEGLLLKAVKATWLTKKNQARIDDVVDYLMMARDSEEYSGSPTIRSRLDEMVVLLTQ
YTRDGVYGSYFNSDEPSLKDEARMVVLELGGLEDRPSLLVAVMFSLILYIENRMYRTPRSLKKLNVIDEG
WRLLDFKNNKVGAFIEKGYRTARRHTGSYITITQNIVDFDSETASSAARAAWGNSSYKIILKQSAKEFAK
YNQLYPDQYSQLQRDMIKGFGSAKDNWFSSFMLSVEHLTSWHRLFVDPLSRAMYSSDGPDFEFLQQRRRE
GLSIHEAVWELAWKKSGPEMAALERWLHEHEGKTIS
MSNPLDSVTRTVNSLLTALKMPDASSEANDVLGQMRFPQFSRVLPYRDYDAETGLFMNEKTMGFMLEAQP
INGANETIVSSLENLMRTKLPREVPVAVHLVSSHLVGQDIEYGLRELSWTGEKADTFNAITRAYYLRAAE
EGFPLPDHLNLPLTLRNYRVYISCCVPRKRGSKASVMEMENQVKIIRASLQGAQIATRSVDADAFIRIAG
EMINHNPDALYPTERELDPLKDLNYQCVDDTFDLKVKADYMTLGLRGNGRNSTARIMNFQLTKNPELAFL
WNMADNYSNLLNPELSVSCPFILTLVLSVEDQVKAQNEANLKFMDLEKKSKTSYAKFFPGVAQEAKEWGD
LRQRLSSGQSSLVSYFLNVTLFCRDDQETALKVEQDVLNSFRKNGFELISPRFNHMRNFLVNLPFMAGGG
LFKQLRESGVLQRAESFNVANLMPVVADSPLATSGLLAPTYRNQLAFIDLFSRGMGNTNFNMAVCGTSGA
GKTGLIQPLIRSVIDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLKFNPFANVTDDTIDEMAERLRDQL
SVMASPNGNLDEVHEGLLLKAVKATWLTKKNQARIDDVVDYLMMARDSEEYSGSPTIRSRLDEMVVLLTQ
YTRDGVYGSYFNSDEPSLKDEARMVVLELGGLEDRPSLLVAVMFSLILYIENRMYRTPRSLKKLNVIDEG
WRLLDFKNNKVGAFIEKGYRTARRHTGSYITITQNIVDFDSETASSAARAAWGNSSYKIILKQSAKEFAK
YNQLYPDQYSQLQRDMIKGFGSAKDNWFSSFMLSVEHLTSWHRLFVDPLSRAMYSSDGPDFEFLQQRRRE
GLSIHEAVWELAWKKSGPEMAALERWLHEHEGKTIS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 11342 | GenBank | WP_077022305 |
| Name | traD_RCR42_RS25235_pB-CRECL423 |
UniProt ID | _ |
| Length | 742 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 742 a.a. Molecular weight: 83244.88 Da Isoelectric Point: 5.1609
>WP_077022305.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIASMRIRMFSQIANIIVYCLVILFGIIVAITLSVKLTWQTFVNGMVYYWCQTLAPMR
DIFKSQPVYTIRYYGHELKYNAQQILQDKYIQWCADQVWTAFILAAIIAAVVCVITFFVTTWILGHQGKQ
QSEDEITGGRQLTDDPKAVAKMLRRDGLASDIKIGELPLIKDSEIQNFCLHGTVGAGKSEVIRRLMNYAR
QRGDMVIVYDRSCEFVKSYYDPATDKILNPLDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLKNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIENNGESFTIRDWMRGVREDGKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDVQTLPDLSCYVTLPGPYPAVKMAL
KYQQRPKVAPEFIPRTIDGKAEERLNAALAAREHESRSIAMLFEPDADVVAPEGGAENAGQPQQPQQPQQ
PQQPQQPQQPQQPQQPQQPQQLTPAPRAADAASVAGAAGAGGVEPELKTKAEEAEQLPPGISESGEVVDM
AAWEAWQAEQDELSPQERQRREEVNINVHRAPEKDLEPGGYI
MSFNAKDMTQGGQIASMRIRMFSQIANIIVYCLVILFGIIVAITLSVKLTWQTFVNGMVYYWCQTLAPMR
DIFKSQPVYTIRYYGHELKYNAQQILQDKYIQWCADQVWTAFILAAIIAAVVCVITFFVTTWILGHQGKQ
QSEDEITGGRQLTDDPKAVAKMLRRDGLASDIKIGELPLIKDSEIQNFCLHGTVGAGKSEVIRRLMNYAR
QRGDMVIVYDRSCEFVKSYYDPATDKILNPLDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLKNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIENNGESFTIRDWMRGVREDGKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDVQTLPDLSCYVTLPGPYPAVKMAL
KYQQRPKVAPEFIPRTIDGKAEERLNAALAAREHESRSIAMLFEPDADVVAPEGGAENAGQPQQPQQPQQ
PQQPQQPQQPQQPQQPQQPQQLTPAPRAADAASVAGAAGAGGVEPELKTKAEEAEQLPPGISESGEVVDM
AAWEAWQAEQDELSPQERQRREEVNINVHRAPEKDLEPGGYI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 82776..107929
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| RCR42_RS25045 (RCR42_25045) | 77873..78196 | + | 324 | WP_008324183 | hypothetical protein | - |
| RCR42_RS25050 (RCR42_25050) | 78258..78617 | + | 360 | WP_015493069 | hypothetical protein | - |
| RCR42_RS25055 (RCR42_25055) | 79244..79618 | + | 375 | WP_008324180 | hypothetical protein | - |
| RCR42_RS25060 (RCR42_25060) | 79672..79959 | + | 288 | WP_008324178 | hypothetical protein | - |
| RCR42_RS25065 (RCR42_25065) | 80006..80350 | + | 345 | WP_008324177 | hypothetical protein | - |
| RCR42_RS25070 (RCR42_25070) | 80436..80699 | + | 264 | WP_008324174 | hypothetical protein | - |
| RCR42_RS25075 (RCR42_25075) | 80742..81572 | + | 831 | WP_008324171 | N-6 DNA methylase | - |
| RCR42_RS25080 (RCR42_25080) | 82199..82729 | + | 531 | WP_008324170 | antirestriction protein | - |
| RCR42_RS25085 (RCR42_25085) | 82776..83252 | - | 477 | WP_008324168 | transglycosylase SLT domain-containing protein | virB1 |
| RCR42_RS25090 (RCR42_25090) | 83684..84076 | + | 393 | WP_008324167 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| RCR42_RS25095 (RCR42_25095) | 84265..84954 | + | 690 | WP_008324166 | hypothetical protein | - |
| RCR42_RS25100 (RCR42_25100) | 85041..85265 | + | 225 | WP_008324165 | TraY domain-containing protein | - |
| RCR42_RS25105 (RCR42_25105) | 85323..85673 | + | 351 | WP_016247536 | type IV conjugative transfer system pilin TraA | - |
| RCR42_RS25110 (RCR42_25110) | 85824..86129 | + | 306 | WP_016479987 | type IV conjugative transfer system protein TraL | traL |
| RCR42_RS25115 (RCR42_25115) | 86144..86710 | + | 567 | WP_016247535 | type IV conjugative transfer system protein TraE | traE |
| RCR42_RS25120 (RCR42_25120) | 86697..87440 | + | 744 | WP_008324157 | type-F conjugative transfer system secretin TraK | traK |
| RCR42_RS25125 (RCR42_25125) | 87427..88821 | + | 1395 | WP_016479986 | F-type conjugal transfer pilus assembly protein TraB | traB |
| RCR42_RS25130 (RCR42_25130) | 88843..89382 | + | 540 | WP_016247533 | type IV conjugative transfer system lipoprotein TraV | traV |
| RCR42_RS25135 (RCR42_25135) | 89502..89726 | + | 225 | WP_008324141 | TraR/DksA C4-type zinc finger protein | - |
| RCR42_RS25140 (RCR42_25140) | 89730..89915 | + | 186 | WP_008324139 | hypothetical protein | - |
| RCR42_RS25145 (RCR42_25145) | 89899..90300 | + | 402 | WP_016479985 | hypothetical protein | - |
| RCR42_RS25150 (RCR42_25150) | 90290..90856 | + | 567 | WP_008324135 | conjugal transfer pilus-stabilizing protein TraP | - |
| RCR42_RS25155 (RCR42_25155) | 90849..93479 | + | 2631 | WP_008324134 | type IV secretion system protein TraC | virb4 |
| RCR42_RS25160 (RCR42_25160) | 93476..93823 | + | 348 | WP_008324133 | type-F conjugative transfer system protein TrbI | - |
| RCR42_RS25165 (RCR42_25165) | 93823..94470 | + | 648 | WP_008324132 | type-F conjugative transfer system protein TraW | traW |
| RCR42_RS25170 (RCR42_25170) | 94473..95459 | + | 987 | WP_042934586 | conjugal transfer pilus assembly protein TraU | traU |
| RCR42_RS25175 (RCR42_25175) | 95476..96105 | + | 630 | WP_008324130 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| RCR42_RS25180 (RCR42_25180) | 96102..97955 | + | 1854 | WP_016479983 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| RCR42_RS25185 (RCR42_25185) | 97987..98175 | + | 189 | WP_008324128 | conjugal transfer protein TrbE | - |
| RCR42_RS25190 (RCR42_25190) | 98206..98955 | + | 750 | WP_008324127 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| RCR42_RS25195 (RCR42_25195) | 98972..99220 | + | 249 | WP_016479982 | type-F conjugative transfer system pilin chaperone TraQ | - |
| RCR42_RS25200 (RCR42_25200) | 99210..99764 | + | 555 | WP_008324125 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| RCR42_RS25205 (RCR42_25205) | 99757..100185 | + | 429 | WP_008324122 | hypothetical protein | - |
| RCR42_RS25210 (RCR42_25210) | 100172..101542 | + | 1371 | WP_016247527 | conjugal transfer pilus assembly protein TraH | traH |
| RCR42_RS25215 (RCR42_25215) | 101542..104364 | + | 2823 | WP_008324119 | conjugal transfer mating-pair stabilization protein TraG | traG |
| RCR42_RS25220 (RCR42_25220) | 104386..104817 | + | 432 | WP_008324118 | entry exclusion protein | - |
| RCR42_RS25225 (RCR42_25225) | 104892..105230 | + | 339 | WP_032072912 | tetratricopeptide repeat protein | - |
| RCR42_RS25230 (RCR42_25230) | 105238..105567 | + | 330 | WP_223856629 | tetratricopeptide repeat protein | - |
| RCR42_RS25235 (RCR42_25235) | 105701..107929 | + | 2229 | WP_077022305 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 15654 | GenBank | NZ_CP133338 |
| Plasmid name | pB-CRECL423 | Incompatibility group | IncFIB |
| Plasmid size | 148248 bp | Coordinate of oriT [Strand] | 83324..83426 [+] |
| Host baterium | Enterobacter hormaechei strain CRECL423 |
Cargo genes
| Drug resistance gene | floR, sul1, rmtC, blaNDM-1 |
| Virulence gene | htpB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |