Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 115202 |
| Name | oriT_pRsp_Pop5_3 |
| Organism | Rhizobium sp. Pop5 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP099398 (214919..214947 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pRsp_Pop5_3
AGGGCGCAATATACGTCGCTGTCGCGACG
AGGGCGCAATATACGTCGCTGTCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file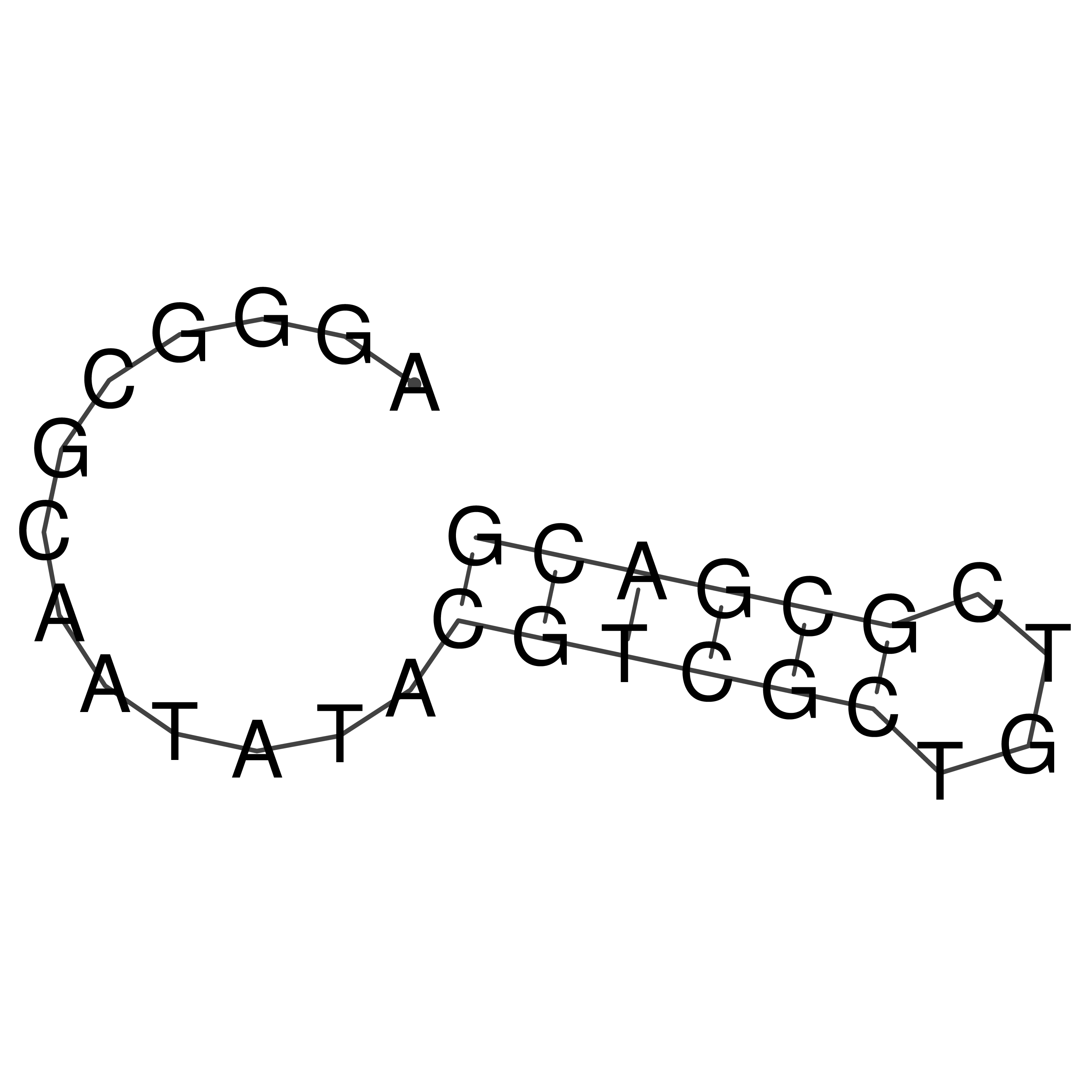
Relaxase
| ID | 9715 | GenBank | WP_258155646 |
| Name | traA_NE852_RS01075_pRsp_Pop5_3 |
UniProt ID | _ |
| Length | 1562 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1562 a.a. Molecular weight: 172328.63 Da Isoelectric Point: 8.0075
>WP_258155646.1 Ti-type conjugative transfer relaxase TraA [Rhizobium sp. Pop5]
MAIMFVRAQVISRGSGRSIVSAAAYRHRARMMDEQAGTSFSYRGGAGELMYEELALPDEIPDWLRSAISG
QSVSKASEVFWNAVDAFETRADAQLARELIIALPEELTRAENITLVREFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGAKKVPVLGEGGKPLRVVTPDRPNGKIVYKVWAGDKETMKAWKIAWAETANR
HLALAGHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGRELHFAPADLARRQEMADRLLSEPELLLKQ
LGNERSTFDERDIARALHRYVDDPTDFANIRARLMASDQLVILKPQEIEAETGKVSEPAVFTTREMLRIE
YDMAQSARVLSERRGFGVSERNVTVAIERVESGDPKNPFRLDAEQVDAVRHVTGDGGIAAIVGLAGAGKS
TLLAAARLAWEGEGHRVIGAALAGKAAEGLQDSSGIKSRTLASWELAWANGRDTLHRGDVLVIDEAGMVA
SQQMARVLKIAEEAEVKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQREAWARNASRLFARGE
VEKGLDAYARHGHLVEAGSREETIDRIVSDWAAARREAIERSTSEGGDGRLRGDELLVLAHTNDDVRKLN
EALRSVMTQEGALGESRSFRSERGAREFAAGDRIIFLENARFLEPRAKHSGPQYVKNGMLGTVVSTGDKR
GDPLLSILLDNGRKLVFSEDSYRHVDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRDRVDL
YAAKEDFAAKPEWGRKPRVDHAAGVTGELVETGQAKFRPDDEDADDSPYADVRADDGTVHRLWGVSLPKA
LEEAGIQEGDTVTLRKDGVERVKVQIAVVDEKTGHKHYEEREVDRNVWTASQVETASARQERIARESHRP
ELFNPLVERLSRSGAKTTTLDFESEASYRAHANDFARRRGLDHLSLAAAEMEQSLTRRWAWIAAKREQVE
KLWERASVALGFAIERERRVAYNEERSQTMVEATSSDARTASHSVSGASAAETRYLIPPATSFVSSVEED
ARLAQLASPAWTEREVILRPLLQKIYRDPDAALVSLNALASDIGVAPRRLADDLAAAPGRLGRLRGSELI
VDGGAAREERNLAVAAVKELLPMARAHATEFRRNAERFVLREQTRRAHMSLSIPALSERAMARLMEIEAV
RRQGGDDAYKTAFALTAKDRSVVQEIKAVSEALTARFGWSAFSAKADAIAERNIVERMPEDLTDERRGKL
TRLFDAVRRFADEQHLAERRDRSKVVAGASVVRGQETENGPGKENVAVPPMLAAVIAFKTSVDDEARLRA
LSNPFYRQQRGALANAATTIWRDPAEVVGKIEELLQKGFAAERIGAAVTNNPAAYGALRGSDRLMDRMLT
SGRERKEAVAAVPEAAARLRALGAAHLNALDAERQAITDERRRMAVAIPALSKAAEEALAHLTVEVSKDS
RKLSVSAASLDPGIGREFAAVSRALDERFGRNALVRGDKDLVNRVPPAQRRAFEAMQERLKVLQQTVRLQ
SSEQIIAERRQRAASRARGINL
MAIMFVRAQVISRGSGRSIVSAAAYRHRARMMDEQAGTSFSYRGGAGELMYEELALPDEIPDWLRSAISG
QSVSKASEVFWNAVDAFETRADAQLARELIIALPEELTRAENITLVREFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGAKKVPVLGEGGKPLRVVTPDRPNGKIVYKVWAGDKETMKAWKIAWAETANR
HLALAGHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGRELHFAPADLARRQEMADRLLSEPELLLKQ
LGNERSTFDERDIARALHRYVDDPTDFANIRARLMASDQLVILKPQEIEAETGKVSEPAVFTTREMLRIE
YDMAQSARVLSERRGFGVSERNVTVAIERVESGDPKNPFRLDAEQVDAVRHVTGDGGIAAIVGLAGAGKS
TLLAAARLAWEGEGHRVIGAALAGKAAEGLQDSSGIKSRTLASWELAWANGRDTLHRGDVLVIDEAGMVA
SQQMARVLKIAEEAEVKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQREAWARNASRLFARGE
VEKGLDAYARHGHLVEAGSREETIDRIVSDWAAARREAIERSTSEGGDGRLRGDELLVLAHTNDDVRKLN
EALRSVMTQEGALGESRSFRSERGAREFAAGDRIIFLENARFLEPRAKHSGPQYVKNGMLGTVVSTGDKR
GDPLLSILLDNGRKLVFSEDSYRHVDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRDRVDL
YAAKEDFAAKPEWGRKPRVDHAAGVTGELVETGQAKFRPDDEDADDSPYADVRADDGTVHRLWGVSLPKA
LEEAGIQEGDTVTLRKDGVERVKVQIAVVDEKTGHKHYEEREVDRNVWTASQVETASARQERIARESHRP
ELFNPLVERLSRSGAKTTTLDFESEASYRAHANDFARRRGLDHLSLAAAEMEQSLTRRWAWIAAKREQVE
KLWERASVALGFAIERERRVAYNEERSQTMVEATSSDARTASHSVSGASAAETRYLIPPATSFVSSVEED
ARLAQLASPAWTEREVILRPLLQKIYRDPDAALVSLNALASDIGVAPRRLADDLAAAPGRLGRLRGSELI
VDGGAAREERNLAVAAVKELLPMARAHATEFRRNAERFVLREQTRRAHMSLSIPALSERAMARLMEIEAV
RRQGGDDAYKTAFALTAKDRSVVQEIKAVSEALTARFGWSAFSAKADAIAERNIVERMPEDLTDERRGKL
TRLFDAVRRFADEQHLAERRDRSKVVAGASVVRGQETENGPGKENVAVPPMLAAVIAFKTSVDDEARLRA
LSNPFYRQQRGALANAATTIWRDPAEVVGKIEELLQKGFAAERIGAAVTNNPAAYGALRGSDRLMDRMLT
SGRERKEAVAAVPEAAARLRALGAAHLNALDAERQAITDERRRMAVAIPALSKAAEEALAHLTVEVSKDS
RKLSVSAASLDPGIGREFAAVSRALDERFGRNALVRGDKDLVNRVPPAQRRAFEAMQERLKVLQQTVRLQ
SSEQIIAERRQRAASRARGINL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 11332 | GenBank | WP_008536019 |
| Name | traG_NE852_RS01060_pRsp_Pop5_3 |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70668.81 Da Isoelectric Point: 9.2416
>WP_008536019.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MGLRGKPHPSLLLVLVPIAVTTITIYIVGWRWPGLAAGMSGKIEHWFLRAAPVPPLLFGPLAGLLTVWAL
PLHRRRPVAMASLLYFLGVAAFYALREFGRLAPAVQAEVITWDRALSYLDMVAVIAAVAGFMAVAMSARI
SVVVPDEIKRARRGIFGDADWLPMTAAGKLFPPDGEIVVGERYRVDKEIIHALPFDVNDRSTWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPMICLDPSTEVAPMVVEHRTNALGREVMVLDPT
NPIMGFNVLDGIEASKQKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYEGRRS
LRSLRQIVSEPEPSVLAMLRDIQEHSGSAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSGDIVSGKKDVFLNISASILRSYPGIARVIIGSLINAMVQADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNLSAQCGEMTVEVKG
SSRNIGWDTKNNASRRSENVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDQAAK
VNRFVKPVL
MGLRGKPHPSLLLVLVPIAVTTITIYIVGWRWPGLAAGMSGKIEHWFLRAAPVPPLLFGPLAGLLTVWAL
PLHRRRPVAMASLLYFLGVAAFYALREFGRLAPAVQAEVITWDRALSYLDMVAVIAAVAGFMAVAMSARI
SVVVPDEIKRARRGIFGDADWLPMTAAGKLFPPDGEIVVGERYRVDKEIIHALPFDVNDRSTWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPMICLDPSTEVAPMVVEHRTNALGREVMVLDPT
NPIMGFNVLDGIEASKQKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYEGRRS
LRSLRQIVSEPEPSVLAMLRDIQEHSGSAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSGDIVSGKKDVFLNISASILRSYPGIARVIIGSLINAMVQADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNLSAQCGEMTVEVKG
SSRNIGWDTKNNASRRSENVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDQAAK
VNRFVKPVL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 191403..202544
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| NE852_RS00895 (NE852_00895) | 186586..186933 | - | 348 | WP_008536653 | IS66 family insertion sequence element accessory protein TnpB | - |
| NE852_RS00900 (NE852_00900) | 187444..187794 | + | 351 | WP_225897245 | transposase | - |
| NE852_RS00905 (NE852_00905) | 187791..188135 | + | 345 | WP_008536637 | IS66 family insertion sequence element accessory protein TnpB | - |
| NE852_RS00910 (NE852_00910) | 188210..189802 | + | 1593 | WP_008536638 | IS66 family transposase | - |
| NE852_RS00915 (NE852_00915) | 190392..190763 | - | 372 | WP_258155640 | MarR family transcriptional regulator | - |
| NE852_RS00920 (NE852_00920) | 190863..191399 | + | 537 | WP_258155641 | hypothetical protein | - |
| NE852_RS00925 (NE852_00925) | 191403..192071 | + | 669 | WP_258155642 | transglycosylase SLT domain-containing protein | virB1 |
| NE852_RS00930 (NE852_00930) | 192068..192367 | + | 300 | WP_008535987 | TrbC/VirB2 family protein | virB2 |
| NE852_RS00935 (NE852_00935) | 192375..192713 | + | 339 | WP_008535988 | type IV secretion system protein VirB3 | virB3 |
| NE852_RS00940 (NE852_00940) | 192706..195072 | + | 2367 | WP_258155643 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| NE852_RS00945 (NE852_00945) | 195069..195770 | + | 702 | WP_258155644 | P-type DNA transfer protein VirB5 | virB5 |
| NE852_RS00950 (NE852_00950) | 195767..196000 | + | 234 | WP_008537008 | EexN family lipoprotein | - |
| NE852_RS00955 (NE852_00955) | 196003..196353 | + | 351 | Protein_187 | type IV secretion system protein | - |
| NE852_RS00960 (NE852_00960) | 196669..197787 | - | 1119 | WP_008536701 | IS110 family transposase | - |
| NE852_RS00965 (NE852_00965) | 197894..198478 | + | 585 | Protein_189 | type IV secretion system protein | - |
| NE852_RS00970 (NE852_00970) | 198518..198799 | + | 282 | WP_080712063 | hypothetical protein | - |
| NE852_RS00975 (NE852_00975) | 198801..199472 | + | 672 | WP_008535994 | virB8 family protein | virB8 |
| NE852_RS00980 (NE852_00980) | 199469..200326 | + | 858 | WP_037146970 | P-type conjugative transfer protein VirB9 | virB9 |
| NE852_RS00985 (NE852_00985) | 200335..201507 | + | 1173 | WP_037146971 | type IV secretion system protein VirB10 | virB10 |
| NE852_RS00990 (NE852_00990) | 201516..202544 | + | 1029 | WP_258155645 | P-type DNA transfer ATPase VirB11 | virB11 |
| NE852_RS00995 (NE852_00995) | 202588..203166 | - | 579 | WP_008536010 | PIN domain-containing protein | - |
| NE852_RS01000 (NE852_01000) | 203156..203677 | - | 522 | WP_008536011 | excisionase | - |
| NE852_RS01005 (NE852_01005) | 204115..205305 | - | 1191 | WP_037146978 | DUF1173 domain-containing protein | - |
| NE852_RS01010 (NE852_01010) | 205616..205873 | + | 258 | Protein_198 | toprim domain-containing protein | - |
| NE852_RS01015 (NE852_01015) | 206111..206973 | + | 863 | Protein_199 | DUF2493 domain-containing protein | - |
| NE852_RS01020 (NE852_01020) | 207058..207324 | + | 267 | WP_004676093 | type II toxin-antitoxin system Phd/YefM family antitoxin | - |
Host bacterium
| ID | 15637 | GenBank | NZ_CP099398 |
| Plasmid name | pRsp_Pop5_3 | Incompatibility group | - |
| Plasmid size | 454824 bp | Coordinate of oriT [Strand] | 214919..214947 [+] |
| Host baterium | Rhizobium sp. Pop5 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | hsiC1/vipB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nifX, nifN, nifE, nifD, nifH, nifS, nifW, fixA, fixB, fixC, fixX, nifA, nifB, nifZ, nifT, nodJ, nodI, nodS, nodC, nodB, fixO, fixN, mLTONO_5203, nifK, nodA, nodD |
| Anti-CRISPR | - |