Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 115195 |
| Name | oriT_pTH2b |
| Organism | Rhizobium sp. TH2 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP062233 (14996..15024 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pTH2b
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file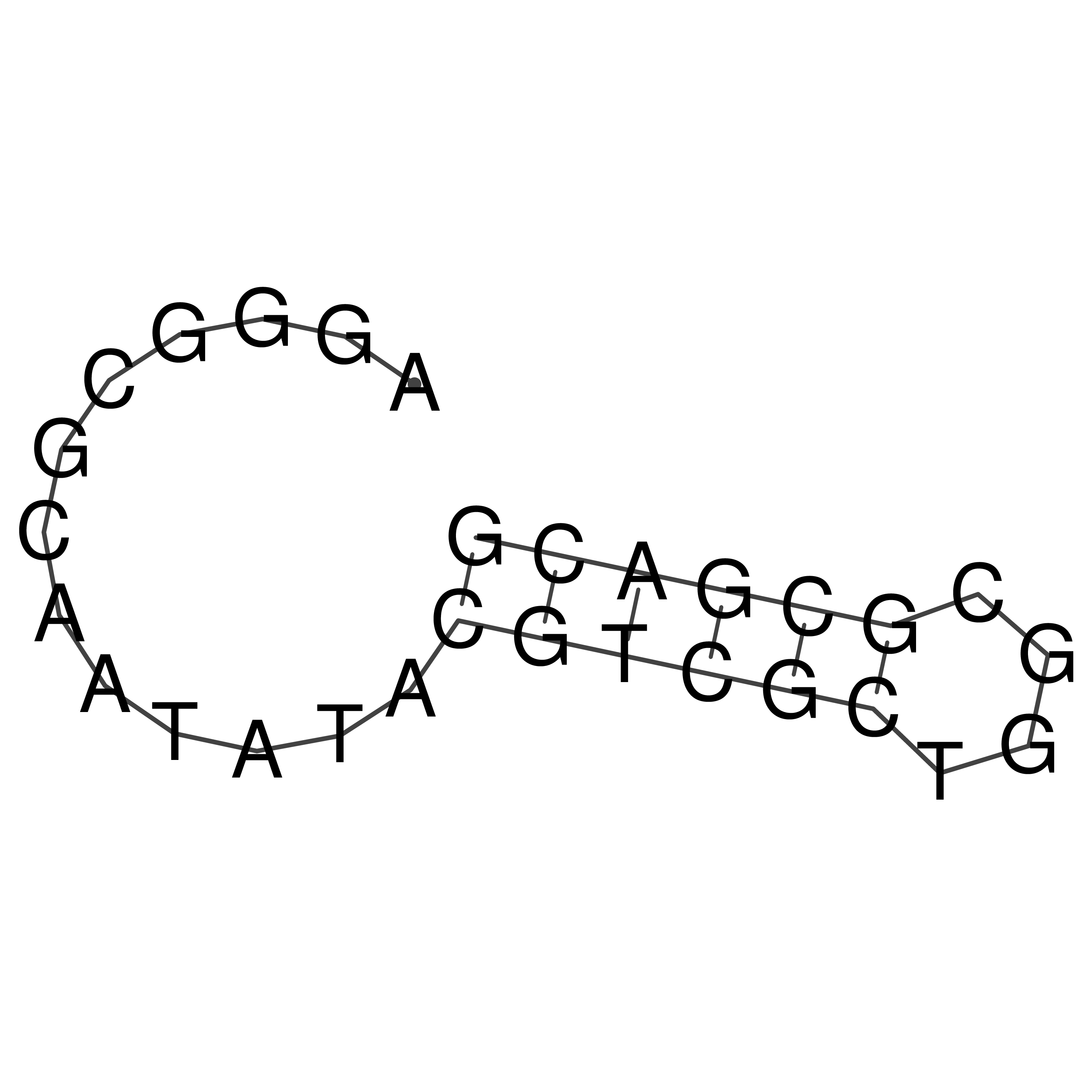
Relaxase
| ID | 9712 | GenBank | WP_258163394 |
| Name | traA_IHQ71_RS30060_pTH2b |
UniProt ID | _ |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 170157.50 Da Isoelectric Point: 8.8948
>WP_258163394.1 Ti-type conjugative transfer relaxase TraA [Rhizobium sp. TH2]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDDQAGTSFSYRGGASELVHEEMALPDQVPGWLKTAIDG
QSVAKASEALWNALDAFETRVDAQLARELIIALPEELTRAENIALVRAFVRDNFTSKGMVADWVYHDKDG
NPHIHLMTTLRPLTEEGFGPKKVPVLGEDGEPLRVATPDRPNGKIVYKLWAGDKETMKAWKIAWAETANQ
HLALAGHEIRLDGRSYAEQGLDRIAQKHLGPEKAALARKGVEMYFAPADLARRQEMADRLLSEPELLLKQ
LGNERSTFDEKDIARALHRYVDDPVDFANIRARLMASDDLVMLRPQQLNAETGKASEPAVFTTREILRIE
YDMARSAEILSERKGFGVSGAQVAAAVKSVETADPANPFKLDAEQVDAVSHVTGNNAIAAVVGLAGAGKS
TLLAAARVAWESEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWASGRETLQRGDVLVVDEAGMVS
SHQMARVLKAVEGVGAKAVLVGDAMQLQPIQAGAAFRAISERVGFAELAGVRRQRDEWAREASRLFARGK
VEEGLDAYAQHGRIAAAETRAEIVSRIVADWTDARRDLLQKTAGGGEASRLRGDELLVLAHTNEDVKRLN
ESLRKVMMEEGALTGAREFQTARGVREFAAGDRIIFLENARFLEPRARRLGPQYVKNGMLGTVVTTGDQR
GDALLSVRLDNGRDVVISEATYRNVDHGYAATIHKSQGSTVDRTFVLATGIMDQHLAYVAMTRHRDRADL
YAAKEDFETKTEWDRKPRADHAAGVTGALVETGFAKFRPNDEDADDSPYADVKTDDGTVHRLWGVSLPKA
IEDAGVADGDTVTLRKNGVEKVKVQIPIIDEQTGQKRFEEREVERNVWTAKQVETVEARQERIERGSHRP
ELFKQLVERLSRSGAKTTTLDFESAAGYQAHAQDFARRRGIDTLASAAAGMEEGVSRRLAWLAEKKEQVA
ELWQRASVALGFAIERERRISYSEDRAESVSAEIPTGRRYLIPPTTDFARSVDEDARRAQLSSPRWNERE
EILRPLLAKIYRDPDVALTHLNALASDARIEPRTLAGDLANAPDRLGRLRGSDNIVEARAARDERSVAVA
AFKELLPLIRAHATEFRRNADRFREREQTRRAYMSLSIPAISKQAMQRLVEIEAVRSRGGDDAWKTAFAF
ATEDRSLVQEVKAVSDALTARFGWSAFTGKADAIAERNVLERMPEDLTDERRGKLTRLFEVIRRFSEEQH
LAERKDQSKIVAGASIEAGKEPNAVLPMLAAVTEFRTPIDDEARPRALATQHYRHNRAALAETATRIWRD
PAGAVDRIEGLIVKGFAAERIAAAVANDPTAYGALRGSDRLMDRMLAVGRERKDALQAVPEASVRLRSLG
VAYVSAFDAERRSVTEERRRMAVAIPGLSQAAEDALRKLAAQMKKQGNKLAVAAGSIDPRIGQEFAAVSR
ALDERFGRNAILRGEKDVVNRVPAAQRRAFEAMHERLMILQQTVRMQASQQIISERQQHIINRARGVIF
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDDQAGTSFSYRGGASELVHEEMALPDQVPGWLKTAIDG
QSVAKASEALWNALDAFETRVDAQLARELIIALPEELTRAENIALVRAFVRDNFTSKGMVADWVYHDKDG
NPHIHLMTTLRPLTEEGFGPKKVPVLGEDGEPLRVATPDRPNGKIVYKLWAGDKETMKAWKIAWAETANQ
HLALAGHEIRLDGRSYAEQGLDRIAQKHLGPEKAALARKGVEMYFAPADLARRQEMADRLLSEPELLLKQ
LGNERSTFDEKDIARALHRYVDDPVDFANIRARLMASDDLVMLRPQQLNAETGKASEPAVFTTREILRIE
YDMARSAEILSERKGFGVSGAQVAAAVKSVETADPANPFKLDAEQVDAVSHVTGNNAIAAVVGLAGAGKS
TLLAAARVAWESEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWASGRETLQRGDVLVVDEAGMVS
SHQMARVLKAVEGVGAKAVLVGDAMQLQPIQAGAAFRAISERVGFAELAGVRRQRDEWAREASRLFARGK
VEEGLDAYAQHGRIAAAETRAEIVSRIVADWTDARRDLLQKTAGGGEASRLRGDELLVLAHTNEDVKRLN
ESLRKVMMEEGALTGAREFQTARGVREFAAGDRIIFLENARFLEPRARRLGPQYVKNGMLGTVVTTGDQR
GDALLSVRLDNGRDVVISEATYRNVDHGYAATIHKSQGSTVDRTFVLATGIMDQHLAYVAMTRHRDRADL
YAAKEDFETKTEWDRKPRADHAAGVTGALVETGFAKFRPNDEDADDSPYADVKTDDGTVHRLWGVSLPKA
IEDAGVADGDTVTLRKNGVEKVKVQIPIIDEQTGQKRFEEREVERNVWTAKQVETVEARQERIERGSHRP
ELFKQLVERLSRSGAKTTTLDFESAAGYQAHAQDFARRRGIDTLASAAAGMEEGVSRRLAWLAEKKEQVA
ELWQRASVALGFAIERERRISYSEDRAESVSAEIPTGRRYLIPPTTDFARSVDEDARRAQLSSPRWNERE
EILRPLLAKIYRDPDVALTHLNALASDARIEPRTLAGDLANAPDRLGRLRGSDNIVEARAARDERSVAVA
AFKELLPLIRAHATEFRRNADRFREREQTRRAYMSLSIPAISKQAMQRLVEIEAVRSRGGDDAWKTAFAF
ATEDRSLVQEVKAVSDALTARFGWSAFTGKADAIAERNVLERMPEDLTDERRGKLTRLFEVIRRFSEEQH
LAERKDQSKIVAGASIEAGKEPNAVLPMLAAVTEFRTPIDDEARPRALATQHYRHNRAALAETATRIWRD
PAGAVDRIEGLIVKGFAAERIAAAVANDPTAYGALRGSDRLMDRMLAVGRERKDALQAVPEASVRLRSLG
VAYVSAFDAERRSVTEERRRMAVAIPGLSQAAEDALRKLAAQMKKQGNKLAVAAGSIDPRIGQEFAAVSR
ALDERFGRNAILRGEKDVVNRVPAAQRRAFEAMHERLMILQQTVRMQASQQIISERQQHIINRARGVIF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 11329 | GenBank | WP_258163392 |
| Name | traG_IHQ71_RS30045_pTH2b |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70352.16 Da Isoelectric Point: 9.1168
>WP_258163392.1 Ti-type conjugative transfer system protein TraG [Rhizobium sp. TH2]
MSGPGKLHPSLLLILVPLTVTAFAAYAVGWRWQELAVGMSGKSAYWFLRAAPVPALLFGPLAGLLSVWAL
PLHRRRPVAMASLVCFLAVAGIYALREFGRLAPSVESGTVTWNQALSYLDFVAVIGALMGFMAVAVTARI
STVVPEPVTRAKRGTFGDADWLSMSAAGKLFPPDGEIAVGERYRVDKEIVHELPFDPNDPATWGQGGRAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYSAPLICLDPSTEVAPMVVEHRTRALGREVMVLDPT
NPIMGFNVLDGIETSKQKEEDIVGIAHMLLSESARFESSTGSYFQNQAHNLLTGLLAHVMLSPEYEGRRT
LRSLRQIVSEPEPSVLAMLRDIQEHSASTFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLESYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMVQADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNIGWDTKNNASRKSESVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMKKEAK
TNRFVKPIP
MSGPGKLHPSLLLILVPLTVTAFAAYAVGWRWQELAVGMSGKSAYWFLRAAPVPALLFGPLAGLLSVWAL
PLHRRRPVAMASLVCFLAVAGIYALREFGRLAPSVESGTVTWNQALSYLDFVAVIGALMGFMAVAVTARI
STVVPEPVTRAKRGTFGDADWLSMSAAGKLFPPDGEIAVGERYRVDKEIVHELPFDPNDPATWGQGGRAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYSAPLICLDPSTEVAPMVVEHRTRALGREVMVLDPT
NPIMGFNVLDGIETSKQKEEDIVGIAHMLLSESARFESSTGSYFQNQAHNLLTGLLAHVMLSPEYEGRRT
LRSLRQIVSEPEPSVLAMLRDIQEHSASTFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLESYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMVQADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNIGWDTKNNASRKSESVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMKKEAK
TNRFVKPIP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 212311..221873
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| IHQ71_RS31255 (IHQ71_31110) | 208523..208741 | + | 219 | WP_258163366 | hypothetical protein | - |
| IHQ71_RS31260 (IHQ71_31115) | 209034..209966 | + | 933 | WP_258163367 | recombinase family protein | - |
| IHQ71_RS31265 (IHQ71_31120) | 210085..210444 | + | 360 | WP_258163368 | VOC family protein | - |
| IHQ71_RS31270 (IHQ71_31125) | 210549..210815 | - | 267 | WP_258163369 | hypothetical protein | - |
| IHQ71_RS31275 (IHQ71_31130) | 210924..211130 | - | 207 | WP_068389465 | cold-shock protein | - |
| IHQ71_RS31280 (IHQ71_31135) | 211458..211676 | + | 219 | WP_258163370 | hypothetical protein | - |
| IHQ71_RS31285 (IHQ71_31140) | 212311..213300 | - | 990 | WP_258163371 | P-type DNA transfer ATPase VirB11 | virB11 |
| IHQ71_RS31290 (IHQ71_31145) | 213309..214475 | - | 1167 | WP_258163372 | type IV secretion system protein VirB10 | virB10 |
| IHQ71_RS31295 (IHQ71_31150) | 214485..215339 | - | 855 | WP_258163373 | P-type conjugative transfer protein VirB9 | virB9 |
| IHQ71_RS31300 (IHQ71_31155) | 215336..216007 | - | 672 | WP_258163374 | virB8 family protein | virB8 |
| IHQ71_RS31305 (IHQ71_31160) | 216013..216285 | - | 273 | WP_258163375 | hypothetical protein | - |
| IHQ71_RS31310 (IHQ71_31165) | 216325..217257 | - | 933 | WP_258163376 | type IV secretion system protein | virB6 |
| IHQ71_RS31315 (IHQ71_31170) | 217261..217494 | - | 234 | WP_258163377 | EexN family lipoprotein | - |
| IHQ71_RS31320 (IHQ71_31175) | 217491..218189 | - | 699 | WP_258163378 | P-type DNA transfer protein VirB5 | virB5 |
| IHQ71_RS31325 (IHQ71_31180) | 218189..220903 | - | 2715 | WP_258163379 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| IHQ71_RS31330 (IHQ71_31185) | 220906..221205 | - | 300 | WP_258163380 | TrbC/VirB2 family protein | virB2 |
| IHQ71_RS31335 (IHQ71_31190) | 221202..221873 | - | 672 | WP_258163381 | lytic transglycosylase domain-containing protein | virB1 |
| IHQ71_RS31340 (IHQ71_31195) | 221876..222397 | - | 522 | WP_258163382 | hypothetical protein | - |
| IHQ71_RS31345 (IHQ71_31200) | 222492..222836 | + | 345 | WP_258163383 | MarR family transcriptional regulator | - |
Host bacterium
| ID | 15630 | GenBank | NZ_CP062233 |
| Plasmid name | pTH2b | Incompatibility group | - |
| Plasmid size | 223174 bp | Coordinate of oriT [Strand] | 14996..15024 [+] |
| Host baterium | Rhizobium sp. TH2 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |