Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 115039 |
| Name | oriT_R-707-1|p1 |
| Organism | Lactococcus lactis strain R-707-1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP102523 (63340..63377 [-], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 1..7, 19..25 (ACACCAC..GTGGTGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 38 nt
>oriT_R-707-1|p1
ACACCACCCAATTTTGGAGTGGTGTGTAAGTGCGCATT
ACACCACCCAATTTTGGAGTGGTGTGTAAGTGCGCATT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file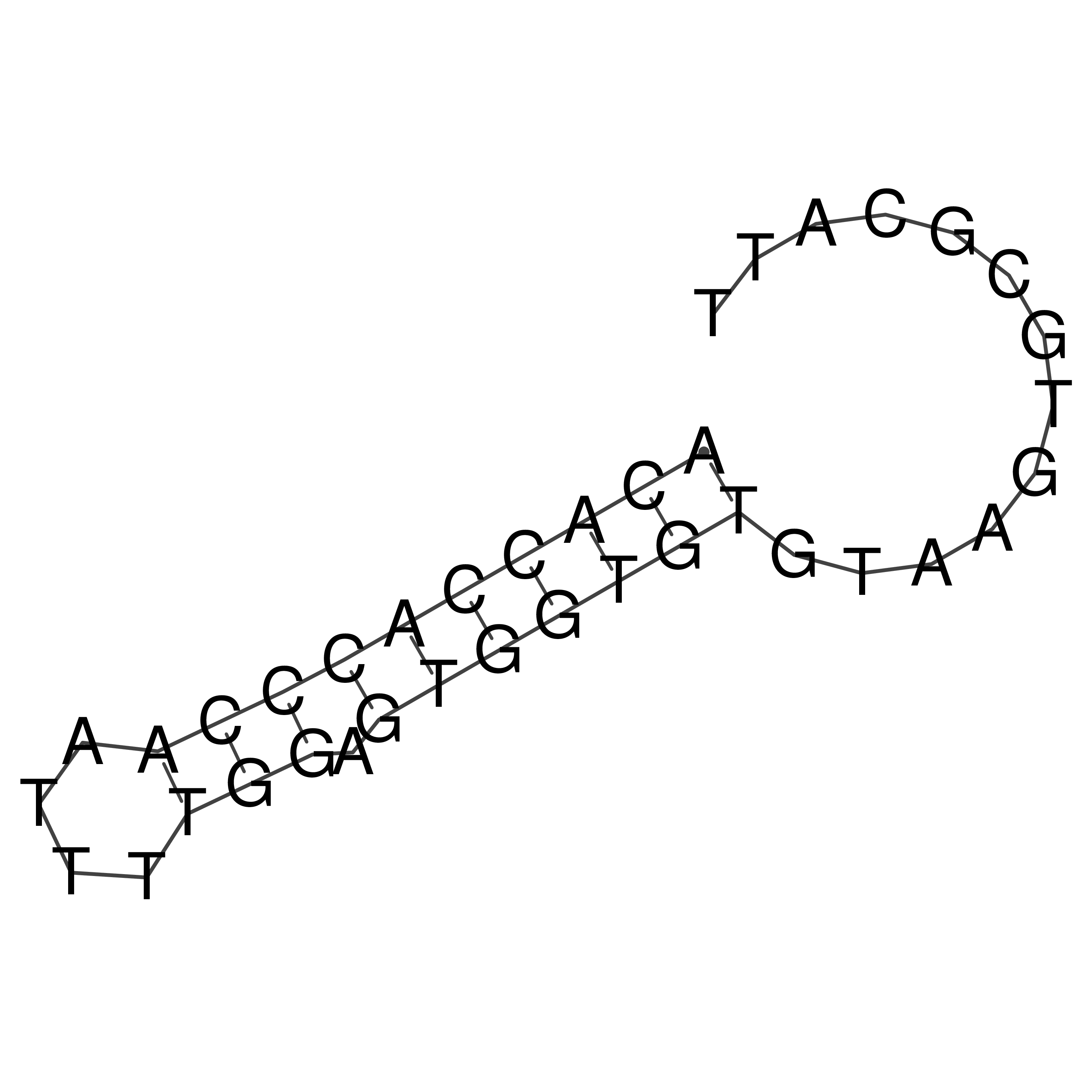
Relaxase
| ID | 9630 | GenBank | WP_010890626 |
| Name | mobQ_NUU08_RS13010_R-707-1|p1 |
UniProt ID | A0A8B3ETK9 |
| Length | 680 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 680 a.a. Molecular weight: 79275.45 Da Isoelectric Point: 6.7052
>WP_010890626.1 MULTISPECIES: MobQ family relaxase [Lactococcus]
MAIFHMNFSNISAGKGRSAVASASYRSGEKLYSEMENKTYFYNRSVMPESFILLPENAPEWAKDRQKLWN
EVEAVDRKVNSRYAKEFNVALPIELSEDEQKELLTEYVQKIFVDKGMVADVAIHRDHDENPHAHVMLTNR
PFNADGSWGQKAKKEYILDENGNKTYTANGHARSRKIWLVDWDKVGKVEEWRKAWADHVNSVFQEKNIDE
RISEKTLEAQGINDIATQHVGVTGNRDERAEFNKLVLENRQHKAELENLDEKINNELKVKQLKNFYSFNE
KKVIAELSKELHTFIDLEHLEEKNKMLFNWKNSVLIKQIVGKDVSEELNKVSRQEFSLDNANRLIDKVVD
RSIKAMYPTVDSNAFSMPEKRQLIRETESENKVFTGAELDDRLSMIRADSLNRQIVALTKRPFTSLTMLS
KQEKYHIDNINNVLAYQGYNYENIEMSHGKILRNYESEEFSKELDIIQRSLKQINSIEEVRKIVEQQYSV
VLGTIFPDSELNKLSLVEKEKTYNAVIYFNPTIKKLTQSELENIVKEPPILFSTKEHNQGLSYLAGRIRL
EDINNKHLTRVLKFEGTKKLFIGEAKADKNIDPELLKEATNRNDKQSYKNEFYRDKNMENYQSVAYSDTS
PAEYMNRLFSGVLISVLYNNENQHKKGLEEVEWDMERKHREHTKTGGLSR
MAIFHMNFSNISAGKGRSAVASASYRSGEKLYSEMENKTYFYNRSVMPESFILLPENAPEWAKDRQKLWN
EVEAVDRKVNSRYAKEFNVALPIELSEDEQKELLTEYVQKIFVDKGMVADVAIHRDHDENPHAHVMLTNR
PFNADGSWGQKAKKEYILDENGNKTYTANGHARSRKIWLVDWDKVGKVEEWRKAWADHVNSVFQEKNIDE
RISEKTLEAQGINDIATQHVGVTGNRDERAEFNKLVLENRQHKAELENLDEKINNELKVKQLKNFYSFNE
KKVIAELSKELHTFIDLEHLEEKNKMLFNWKNSVLIKQIVGKDVSEELNKVSRQEFSLDNANRLIDKVVD
RSIKAMYPTVDSNAFSMPEKRQLIRETESENKVFTGAELDDRLSMIRADSLNRQIVALTKRPFTSLTMLS
KQEKYHIDNINNVLAYQGYNYENIEMSHGKILRNYESEEFSKELDIIQRSLKQINSIEEVRKIVEQQYSV
VLGTIFPDSELNKLSLVEKEKTYNAVIYFNPTIKKLTQSELENIVKEPPILFSTKEHNQGLSYLAGRIRL
EDINNKHLTRVLKFEGTKKLFIGEAKADKNIDPELLKEATNRNDKQSYKNEFYRDKNMENYQSVAYSDTS
PAEYMNRLFSGVLISVLYNNENQHKKGLEEVEWDMERKHREHTKTGGLSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A8B3ETK9 |
T4CP
| ID | 11236 | GenBank | WP_010890635 |
| Name | t4cp2_NUU08_RS13070_R-707-1|p1 |
UniProt ID | _ |
| Length | 530 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 530 a.a. Molecular weight: 59615.30 Da Isoelectric Point: 4.8001
>WP_010890635.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Lactococcus]
MNGTILGVLDNKIIYQDNTTKPNRNVMVIGGSGSYKTQSVVITNLFNETKNSIVVTDPKGELYEKTAGIK
LAQGYEVHVVNFANMAHSDRYNPFDYIERDIQAESVATKIVQSENAEGKKDVWFSTQRQLLKALILFVMK
ERSPEQRNLAGVINVLQTFDSEPINKDENSDLDNLFLALKITHPARIAYELGFKKAKGDMKASIISSLLA
TISKFTDEEVSNFTSISDFHLQDIGRKKIVLYVIIPVMDNTYESFINLFFSQMFDELYKLASSNGAKLPQ
EVDFILDEFVNLGKFPKYEEFLATCRGYGIGVTTICQTLTQLQSLYGKEKAESILGNHAVKICLNASNEA
TAKYFSELLGKSTVKVETGSESTSHSKETSTSKSDSYSYTSRQLMTPDEIIRMPDTQSLLIFTNQKPIKA
TKAFQFKLFPDADSKVKLEQNKYVGITSKSQLEKYNDLSVKWEEKLQSLKNITVTEEEEKDLQDNIDSDL
EAMNNENSGVDNSTTETDKIAETEAKEVQAEEKNFDQVVL
MNGTILGVLDNKIIYQDNTTKPNRNVMVIGGSGSYKTQSVVITNLFNETKNSIVVTDPKGELYEKTAGIK
LAQGYEVHVVNFANMAHSDRYNPFDYIERDIQAESVATKIVQSENAEGKKDVWFSTQRQLLKALILFVMK
ERSPEQRNLAGVINVLQTFDSEPINKDENSDLDNLFLALKITHPARIAYELGFKKAKGDMKASIISSLLA
TISKFTDEEVSNFTSISDFHLQDIGRKKIVLYVIIPVMDNTYESFINLFFSQMFDELYKLASSNGAKLPQ
EVDFILDEFVNLGKFPKYEEFLATCRGYGIGVTTICQTLTQLQSLYGKEKAESILGNHAVKICLNASNEA
TAKYFSELLGKSTVKVETGSESTSHSKETSTSKSDSYSYTSRQLMTPDEIIRMPDTQSLLIFTNQKPIKA
TKAFQFKLFPDADSKVKLEQNKYVGITSKSQLEKYNDLSVKWEEKLQSLKNITVTEEEEKDLQDNIDSDL
EAMNNENSGVDNSTTETDKIAETEAKEVQAEEKNFDQVVL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 58564..73943
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| NUU08_RS12950 (NUU08_12940) | 53738..54295 | + | 558 | WP_015081779 | recombinase family protein | - |
| NUU08_RS12955 (NUU08_12945) | 54439..55236 | + | 798 | WP_015081778 | ParA family protein | - |
| NUU08_RS12960 (NUU08_12950) | 55250..55516 | + | 267 | WP_010890679 | hypothetical protein | - |
| NUU08_RS12965 (NUU08_12955) | 55766..56989 | + | 1224 | WP_003331415 | IS21-like element IS712 family transposase | - |
| NUU08_RS12970 (NUU08_12960) | 57001..57759 | + | 759 | WP_003331414 | IS21-like element IS712 family helper ATPase IstB | - |
| NUU08_RS12975 (NUU08_12965) | 58158..58538 | + | 381 | WP_010890681 | single-stranded DNA-binding protein | - |
| NUU08_RS12980 (NUU08_12970) | 58564..59508 | + | 945 | WP_063283863 | toprim domain-containing protein | traP |
| NUU08_RS12985 (NUU08_12975) | 60180..61334 | + | 1155 | WP_010890657 | RepB family plasmid replication initiator protein | - |
| NUU08_RS12990 (NUU08_12980) | 61813..62130 | + | 318 | WP_058147852 | hypothetical protein | - |
| NUU08_RS12995 (NUU08_12985) | 62145..62345 | + | 201 | WP_058147838 | hypothetical protein | - |
| NUU08_RS13000 (NUU08_12990) | 62628..62894 | - | 267 | WP_010890660 | hypothetical protein | - |
| NUU08_RS13005 (NUU08_12995) | 62907..63191 | - | 285 | WP_058147839 | hypothetical protein | - |
| NUU08_RS13010 (NUU08_13000) | 63460..65502 | + | 2043 | WP_010890626 | MobQ family relaxase | - |
| NUU08_RS13015 (NUU08_13005) | 65588..65932 | + | 345 | WP_010890662 | hypothetical protein | - |
| NUU08_RS13020 (NUU08_13010) | 65933..66538 | + | 606 | WP_010890663 | hypothetical protein | - |
| NUU08_RS13025 (NUU08_13015) | 66551..66895 | + | 345 | WP_058147840 | CagC family type IV secretion system protein | - |
| NUU08_RS13030 (NUU08_13020) | 66916..67263 | + | 348 | WP_010890628 | hypothetical protein | trsC |
| NUU08_RS13035 (NUU08_13025) | 67247..67900 | + | 654 | WP_010890629 | TrsD/TraD family conjugative transfer protein | trsD |
| NUU08_RS13040 (NUU08_13030) | 67912..69930 | + | 2019 | WP_010890630 | VirB4 family type IV secretion system protein | virb4 |
| NUU08_RS13045 (NUU08_13035) | 69923..71341 | + | 1419 | WP_058147841 | type VII secretion protein EssB/YukC | - |
| NUU08_RS13050 (NUU08_13040) | 71343..72500 | + | 1158 | WP_010890632 | phage tail tip lysozyme | prgK |
| NUU08_RS13055 (NUU08_13045) | 72513..73130 | + | 618 | WP_010890664 | hypothetical protein | - |
| NUU08_RS13060 (NUU08_13050) | 73123..73488 | + | 366 | WP_223203862 | thioredoxin | - |
| NUU08_RS13065 (NUU08_13055) | 73485..73943 | + | 459 | WP_010890634 | hypothetical protein | trsJ |
| NUU08_RS13070 (NUU08_13060) | 73940..75532 | + | 1593 | WP_010890635 | VirD4-like conjugal transfer protein, CD1115 family | - |
| NUU08_RS13075 (NUU08_13065) | 75547..75921 | + | 375 | WP_058147842 | hypothetical protein | - |
Host bacterium
| ID | 15474 | GenBank | NZ_CP102523 |
| Plasmid name | R-707-1|p1 | Incompatibility group | - |
| Plasmid size | 75979 bp | Coordinate of oriT [Strand] | 63340..63377 [-] |
| Host baterium | Lactococcus lactis strain R-707-1 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |