Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114805 |
| Name | oriT_253_2_EW_B|plas2 |
| Organism | Escherichia albertii strain 253_2_EW_B |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP099873 (3259..3307 [+], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_253_2_EW_B|plas2
CGATTGTAACATACCCGGAACGGGTTTGTGTACAATCGGTCTCGTGCCT
CGATTGTAACATACCCGGAACGGGTTTGTGTACAATCGGTCTCGTGCCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file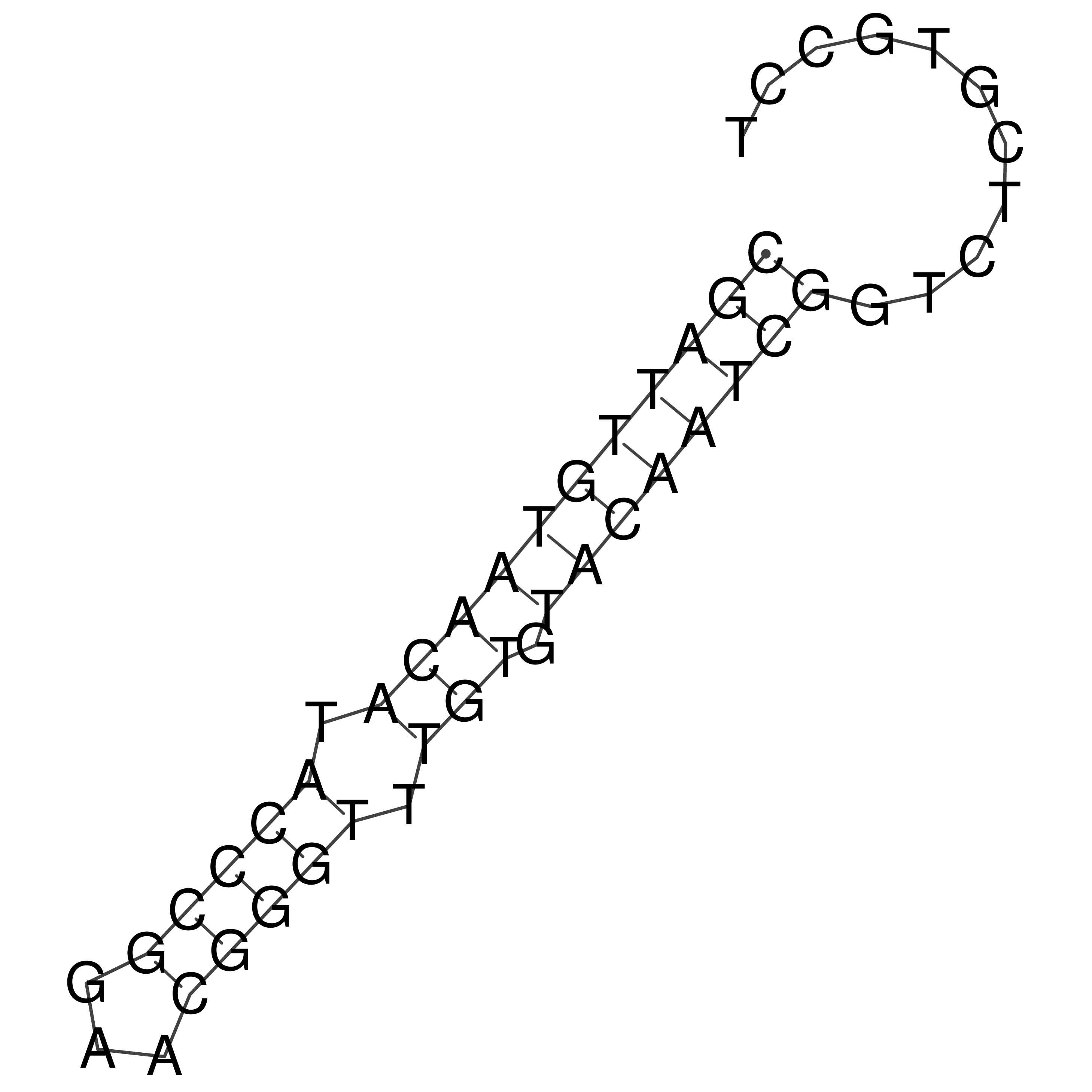
Relaxase
| ID | 9489 | GenBank | WP_306296733 |
| Name | Relaxase_NIZ24_RS24005_253_2_EW_B|plas2 |
UniProt ID | _ |
| Length | 320 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 320 a.a. Molecular weight: 37368.23 Da Isoelectric Point: 9.8860
>WP_306296733.1 relaxase/mobilization nuclease domain-containing protein [Escherichia albertii]
MIIRYGGGNHGIAEYLENGRKAEREYTRDELDHRFIIDGDLATTNKIIESIEDKGQERYLHITLSFAENE
ISEETLKNVTSDFKKMFMNAYHTDEYSFYAEAHLPKIKNIVDNKTGELVERKPHIHIVIPRTNLVTGRSL
NPRGDLTNLKTQTQLDSVQEYLNNKYNLISPKDSVRVSDENYANVLSRVKGDLYRERNNEVKKEVFSRVT
NENINSTDAFKNLLMQYGDVKVRNEGKTNQYFAVRLKDDKKYTNFKNPLFSSSFIEKRELPLVKPTPAQI
NNNVSTWLDKTSHEIKHIYSNRVKRVSTIRRLVSRKRKVF
MIIRYGGGNHGIAEYLENGRKAEREYTRDELDHRFIIDGDLATTNKIIESIEDKGQERYLHITLSFAENE
ISEETLKNVTSDFKKMFMNAYHTDEYSFYAEAHLPKIKNIVDNKTGELVERKPHIHIVIPRTNLVTGRSL
NPRGDLTNLKTQTQLDSVQEYLNNKYNLISPKDSVRVSDENYANVLSRVKGDLYRERNNEVKKEVFSRVT
NENINSTDAFKNLLMQYGDVKVRNEGKTNQYFAVRLKDDKKYTNFKNPLFSSSFIEKRELPLVKPTPAQI
NNNVSTWLDKTSHEIKHIYSNRVKRVSTIRRLVSRKRKVF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 11047 | GenBank | WP_000726942 |
| Name | t4cp2_NIZ24_RS23775_253_2_EW_B|plas2 |
UniProt ID | _ |
| Length | 655 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 655 a.a. Molecular weight: 74097.02 Da Isoelectric Point: 8.2345
>WP_000726942.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Enterobacteriaceae]
MKKKIILVVFLLLILVVCLIGGNYLGGYAALKYSGLNIDMLQWNTFNNVITQFSGKPEYKKLVAITWAGF
AAPCAIFIGFVVIVIVGLMPKKVIYGNARLATDMDLAKSTFFPTDKELKEARLKNSKPYCFPPILIGKQF
KGRFKNKYIYFFGQQFLILYAPTRSGKGVGIVIPNCVNYPDSMVVLDIKLENWFLSAGYRQNVLGQECFL
FAPAGYAENQQEAKKGNIRSHRWNPLDCVNRSDIQRSGDLEKIAAMLIPASDDPIWSDSARKLFCGLALY
LLDKERFHLQQKKKGVADVPDVLVSMSAIMKLSVPESGQKLSAWMGTEIDQKEYLSDETKSLFREFMAAP
EKTQGSIITNFSSPLSIFKNPITAAATNASDFDVRMVRKKPMSIYLGLTPDALITHSRLVNLFFSMLVNE
NTKELPEQNPELKYQCLVLLDEFTSMGKSEIIEKGVGFTAGFNLRFVIILQNEGQGKKDDMYGTHGWETF
VENSAVVLYYPPKAKNELAKKISEEIGVRDMKIIKKSDSRSGGKGGSSRSRNHEIVSRAVLLPEEIVELR
DYKNKMGNMAVREIVMSEYTRPFVANKIIWFEEEEFKKRVDIAKENPVEMPVLFDEEMRNQILEQAKIYS
DDISMFSDVMTKPDLETHDLEDVSE
MKKKIILVVFLLLILVVCLIGGNYLGGYAALKYSGLNIDMLQWNTFNNVITQFSGKPEYKKLVAITWAGF
AAPCAIFIGFVVIVIVGLMPKKVIYGNARLATDMDLAKSTFFPTDKELKEARLKNSKPYCFPPILIGKQF
KGRFKNKYIYFFGQQFLILYAPTRSGKGVGIVIPNCVNYPDSMVVLDIKLENWFLSAGYRQNVLGQECFL
FAPAGYAENQQEAKKGNIRSHRWNPLDCVNRSDIQRSGDLEKIAAMLIPASDDPIWSDSARKLFCGLALY
LLDKERFHLQQKKKGVADVPDVLVSMSAIMKLSVPESGQKLSAWMGTEIDQKEYLSDETKSLFREFMAAP
EKTQGSIITNFSSPLSIFKNPITAAATNASDFDVRMVRKKPMSIYLGLTPDALITHSRLVNLFFSMLVNE
NTKELPEQNPELKYQCLVLLDEFTSMGKSEIIEKGVGFTAGFNLRFVIILQNEGQGKKDDMYGTHGWETF
VENSAVVLYYPPKAKNELAKKISEEIGVRDMKIIKKSDSRSGGKGGSSRSRNHEIVSRAVLLPEEIVELR
DYKNKMGNMAVREIVMSEYTRPFVANKIIWFEEEEFKKRVDIAKENPVEMPVLFDEEMRNQILEQAKIYS
DDISMFSDVMTKPDLETHDLEDVSE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 13915..22688
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| NIZ24_RS23705 (NIZ24_23685) | 10783..11787 | + | 1005 | WP_001208090 | replication initiation protein | - |
| NIZ24_RS23710 (NIZ24_23690) | 12161..12499 | - | 339 | WP_000612441 | helix-turn-helix domain-containing protein | - |
| NIZ24_RS23715 (NIZ24_23695) | 12698..12985 | + | 288 | WP_000332248 | helix-turn-helix transcriptional regulator | - |
| NIZ24_RS23720 (NIZ24_23700) | 13026..13259 | - | 234 | WP_001056371 | EexN family lipoprotein | - |
| NIZ24_RS23725 (NIZ24_23705) | 13272..13913 | - | 642 | WP_000844300 | type IV secretion system protein | - |
| NIZ24_RS23730 (NIZ24_23710) | 13915..14910 | - | 996 | WP_001103652 | type IV secretion system protein | virB6 |
| NIZ24_RS23735 (NIZ24_23715) | 15243..15854 | + | 612 | WP_000603936 | lytic transglycosylase domain-containing protein | virB1 |
| NIZ24_RS23740 (NIZ24_23720) | 15847..16140 | + | 294 | WP_001031682 | TrbC/VirB2 family protein | virB2 |
| NIZ24_RS23745 (NIZ24_23725) | 16198..16518 | + | 321 | WP_001000143 | VirB3 family type IV secretion system protein | virB3 |
| NIZ24_RS23750 (NIZ24_23730) | 16521..18893 | + | 2373 | WP_053293934 | VirB4 family type IV secretion system protein | virb4 |
| NIZ24_RS23755 (NIZ24_23735) | 19024..19761 | + | 738 | WP_000364523 | virB8 family protein | virB8 |
| NIZ24_RS23760 (NIZ24_23740) | 19758..20537 | + | 780 | WP_000592647 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| NIZ24_RS23765 (NIZ24_23745) | 20527..21669 | + | 1143 | WP_032351177 | TrbI/VirB10 family protein | virB10 |
| NIZ24_RS23770 (NIZ24_23750) | 21669..22688 | + | 1020 | WP_001287983 | P-type DNA transfer ATPase VirB11 | virB11 |
| NIZ24_RS23775 (NIZ24_23755) | 22739..24706 | + | 1968 | WP_000726942 | type IV secretory system conjugative DNA transfer family protein | - |
| NIZ24_RS23780 (NIZ24_23760) | 24722..25027 | + | 306 | WP_001223992 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| NIZ24_RS23785 (NIZ24_23765) | 25150..25737 | + | 588 | WP_000280687 | DUF6012 family protein | - |
Host bacterium
| ID | 15240 | GenBank | NZ_CP099873 |
| Plasmid name | 253_2_EW_B|plas2 | Incompatibility group | IncX4 |
| Plasmid size | 37918 bp | Coordinate of oriT [Strand] | 3259..3307 [+] |
| Host baterium | Escherichia albertii strain 253_2_EW_B |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |