Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114800 |
| Name | oriT_147_1_TBG_A|plas2 |
| Organism | Escherichia albertii strain 147_1_TBG_A |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP099900 (5727..5827 [+], 101 nt) |
| oriT length | 101 nt |
| IRs (inverted repeats) | 32..38, 43..49 (TTACGAT..ATCGTAA) 21..26, 38..43 (TTTTCA..TGAAAA) |
| Location of nic site | 62..63 |
| Conserved sequence flanking the nic site |
GGTGTATAGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 101 nt
>oriT_147_1_TBG_A|plas2
AATTAGGATAATTTGTTTTGTTTTCAAGCATTTACGATGAAAATCGTAATTGCGTATGGTGTATAGCCGTTAAGGGATACCATACCACGCCTTTTTTAAGG
AATTAGGATAATTTGTTTTGTTTTCAAGCATTTACGATGAAAATCGTAATTGCGTATGGTGTATAGCCGTTAAGGGATACCATACCACGCCTTTTTTAAGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file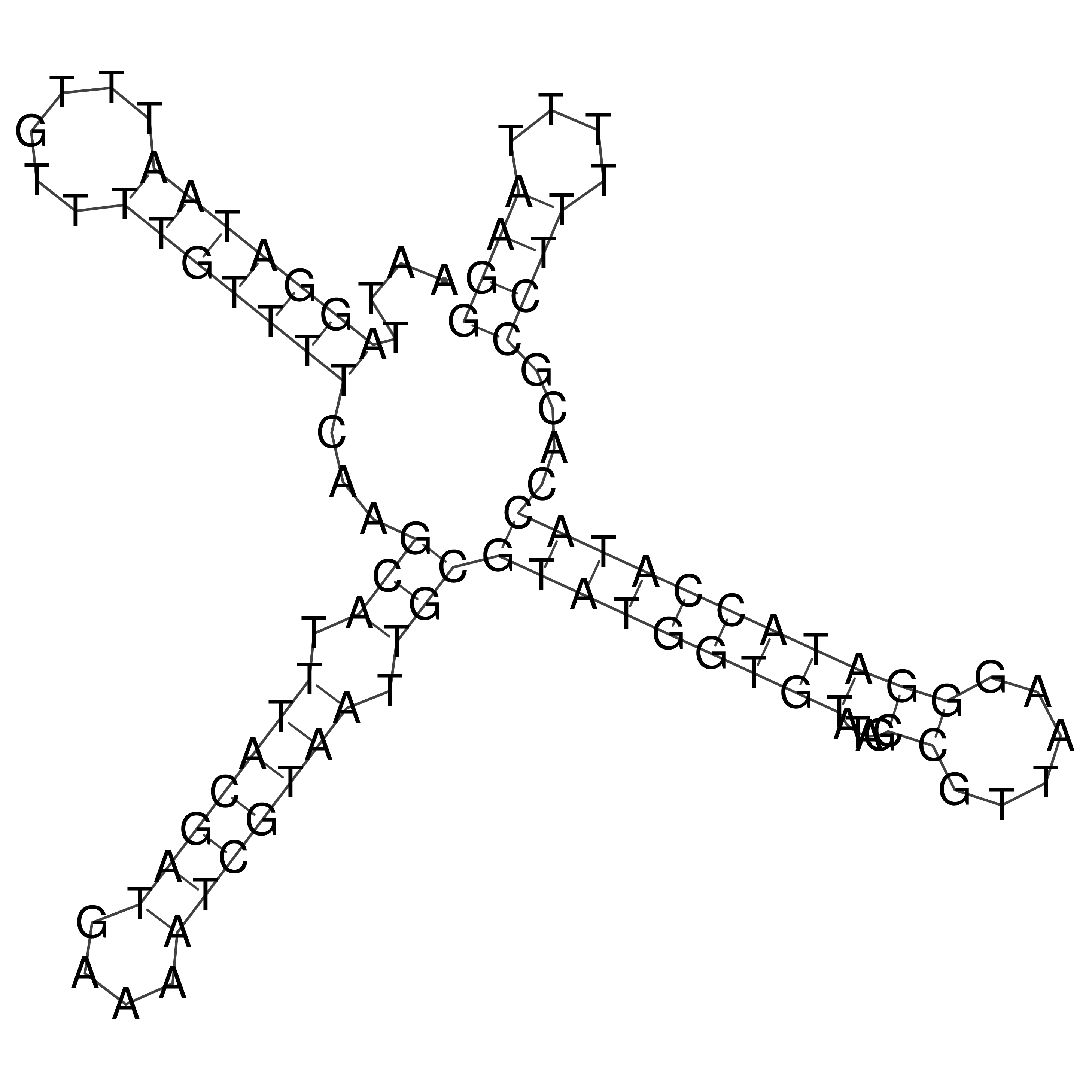
Relaxase
| ID | 9485 | GenBank | WP_256879687 |
| Name | mobF_NIZ13_RS23670_147_1_TBG_A|plas2 |
UniProt ID | _ |
| Length | 295 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 295 a.a. Molecular weight: 33330.35 Da Isoelectric Point: 7.5115
>WP_256879687.1 MobF family relaxase [Escherichia albertii]
MLNITTITRQSACDKVVDYYADGADDYYAKDGNAMQWQGAGAEELGLTGEVEQKRFAKLLDGKVTDDISV
MRKTANSESKERLGYDLTFSAPKGVTLQALVNGDKRIIEAHDRAVTKAIEEAERLALGRFTVNKKTHVEN
TNKLIVGKFRHETSRELDPDLHTHAFVMNLTKTSDGKWRSLTNDGIVNSLTLLGNVYKSELARELELAGY
NLRYDRNGTFDLAHFSQEQIAQFSQRSQQIEAELAKNGLSRETASHEQKNAAALKTRKSKGEVDRDVLYE
GWKNRAKELQIWTWR
MLNITTITRQSACDKVVDYYADGADDYYAKDGNAMQWQGAGAEELGLTGEVEQKRFAKLLDGKVTDDISV
MRKTANSESKERLGYDLTFSAPKGVTLQALVNGDKRIIEAHDRAVTKAIEEAERLALGRFTVNKKTHVEN
TNKLIVGKFRHETSRELDPDLHTHAFVMNLTKTSDGKWRSLTNDGIVNSLTLLGNVYKSELARELELAGY
NLRYDRNGTFDLAHFSQEQIAQFSQRSQQIEAELAKNGLSRETASHEQKNAAALKTRKSKGEVDRDVLYE
GWKNRAKELQIWTWR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 11040 | GenBank | WP_001001035 |
| Name | t4cp2_NIZ13_RS23675_147_1_TBG_A|plas2 |
UniProt ID | _ |
| Length | 525 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 525 a.a. Molecular weight: 59019.93 Da Isoelectric Point: 9.5507
>WP_001001035.1 MULTISPECIES: type IV secretion system DNA-binding domain-containing protein [Enterobacterales]
MNEADKGKICLGALLFTPLVTWFIAAKTLYGFTPKDARERLVLLIQQTFDLWPLWGALLVGEIIAIIGLI
IFFKFNHQIFKGAPFKKIYRGTRLVSQRALASLTKEREPQITIADIPIPTKAEGTHISIAGATGVGKSTI
FAEMMKGCLMRGKMQRVASKKDRMIILDPDGDFLSRFYKKGDKILNPYDARTEGWVFFNEIKSDFDFERF
GVSIVPTAKDSQTEEWNGYGRLLFTEVSRKVFNTSRNPTMEEVFYWTNEVPLEELEEYVRGTKAQALFAG
SGRAVGSARFVLSNKLAAHLKMPAGNFSIREWLEDPKGGNLYITWTDEMREALKSLISCWTDSIFSIVLG
MSKSNTRKIWTYIDELESLDYLPSLRDALTKGRKKGLRVVTGYQSYSQVISIYGSDVAETLLSNHRTSVV
MAAGRLGERTLEFVSKSLGEIEGEREKTGISRRFGQMGTKNINDDHKRERAVTPTEIATLDDLTGYIAFP
GNLPVAKFETHHVNYTRSNPVPGVVLRESTAFAGM
MNEADKGKICLGALLFTPLVTWFIAAKTLYGFTPKDARERLVLLIQQTFDLWPLWGALLVGEIIAIIGLI
IFFKFNHQIFKGAPFKKIYRGTRLVSQRALASLTKEREPQITIADIPIPTKAEGTHISIAGATGVGKSTI
FAEMMKGCLMRGKMQRVASKKDRMIILDPDGDFLSRFYKKGDKILNPYDARTEGWVFFNEIKSDFDFERF
GVSIVPTAKDSQTEEWNGYGRLLFTEVSRKVFNTSRNPTMEEVFYWTNEVPLEELEEYVRGTKAQALFAG
SGRAVGSARFVLSNKLAAHLKMPAGNFSIREWLEDPKGGNLYITWTDEMREALKSLISCWTDSIFSIVLG
MSKSNTRKIWTYIDELESLDYLPSLRDALTKGRKKGLRVVTGYQSYSQVISIYGSDVAETLLSNHRTSVV
MAAGRLGERTLEFVSKSLGEIEGEREKTGISRRFGQMGTKNINDDHKRERAVTPTEIATLDDLTGYIAFP
GNLPVAKFETHHVNYTRSNPVPGVVLRESTAFAGM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 22136..33942
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| NIZ13_RS23785 (NIZ13_23770) | 17788..18372 | - | 585 | WP_000690619 | recombinase family protein | - |
| NIZ13_RS23790 (NIZ13_23775) | 18372..18794 | - | 423 | WP_061090063 | hypothetical protein | - |
| NIZ13_RS23795 (NIZ13_23780) | 19259..19504 | + | 246 | WP_000195442 | AbrB/MazE/SpoVT family DNA-binding domain-containing protein | - |
| NIZ13_RS23800 (NIZ13_23785) | 19504..20046 | + | 543 | WP_000711249 | hypothetical protein | - |
| NIZ13_RS23805 (NIZ13_23790) | 20016..20312 | + | 297 | WP_226331936 | hypothetical protein | - |
| NIZ13_RS23810 (NIZ13_23795) | 20302..20628 | + | 327 | WP_000817647 | endoribonuclease MazF | - |
| NIZ13_RS23815 (NIZ13_23800) | 20654..20968 | + | 315 | WP_001280992 | hypothetical protein | - |
| NIZ13_RS24865 | 21028..21429 | - | 402 | WP_015345245 | Hha/YmoA family nucleoid-associated regulatory protein | - |
| NIZ13_RS23825 (NIZ13_23810) | 21573..22160 | - | 588 | WP_256879679 | restriction endonuclease | - |
| NIZ13_RS23830 (NIZ13_23815) | 22136..22450 | - | 315 | WP_000851068 | hypothetical protein | tfc15 |
| NIZ13_RS23835 (NIZ13_23820) | 22453..22677 | - | 225 | WP_239534999 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| NIZ13_RS23840 (NIZ13_23825) | 22780..23217 | - | 438 | WP_000034608 | hypothetical protein | - |
| NIZ13_RS23845 (NIZ13_23830) | 23229..23465 | - | 237 | WP_000089855 | H-NS family nucleoid-associated regulatory protein | - |
| NIZ13_RS23850 (NIZ13_23835) | 23528..24196 | + | 669 | WP_256879680 | lytic transglycosylase domain-containing protein | virB1 |
| NIZ13_RS23855 (NIZ13_23840) | 24290..24607 | + | 318 | WP_001267114 | hypothetical protein | - |
| NIZ13_RS23860 (NIZ13_23845) | 24698..24880 | + | 183 | WP_256879681 | TrbC/VirB2 family protein | virB2 |
| NIZ13_RS23865 (NIZ13_23850) | 24930..25190 | + | 261 | WP_256879682 | VirB3 family type IV secretion system protein | virB3 |
| NIZ13_RS23870 (NIZ13_23855) | 25242..26684 | + | 1443 | WP_256879683 | hypothetical protein | virb4 |
| NIZ13_RS23875 (NIZ13_23860) | 26700..27872 | + | 1173 | WP_252363867 | AAA family ATPase | virb4 |
| NIZ13_RS23880 (NIZ13_23865) | 27882..28589 | + | 708 | WP_036896352 | type IV secretion system protein | virB5 |
| NIZ13_RS23885 (NIZ13_23870) | 28593..28829 | + | 237 | WP_000733633 | EexN family lipoprotein | - |
| NIZ13_RS23890 (NIZ13_23875) | 29186..29893 | + | 708 | Protein_47 | type IV secretion system protein | - |
| NIZ13_RS23895 (NIZ13_23880) | 29988..30158 | + | 171 | WP_021536809 | hypothetical protein | - |
| NIZ13_RS23900 (NIZ13_23885) | 30161..30877 | + | 717 | WP_000914645 | virB8 family protein | virB8 |
| NIZ13_RS23905 (NIZ13_23890) | 30880..31767 | + | 888 | WP_000744355 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| NIZ13_RS23910 (NIZ13_23895) | 31764..32918 | + | 1155 | WP_001224296 | type IV secretion system protein VirB10 | virB10 |
| NIZ13_RS23915 (NIZ13_23900) | 32911..33942 | + | 1032 | WP_000138004 | P-type DNA transfer ATPase VirB11 | virB11 |
| NIZ13_RS23920 (NIZ13_23905) | 33905..34459 | + | 555 | WP_000616063 | phospholipase D family protein | - |
Host bacterium
| ID | 15235 | GenBank | NZ_CP099900 |
| Plasmid name | 147_1_TBG_A|plas2 | Incompatibility group | - |
| Plasmid size | 34480 bp | Coordinate of oriT [Strand] | 5727..5827 [+] |
| Host baterium | Escherichia albertii strain 147_1_TBG_A |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |