Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114799 |
| Name | oriT_231_1_NP_B|plas2 |
| Organism | Escherichia albertii strain 231_1_NP_B |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP099885 (26737..26860 [+], 124 nt) |
| oriT length | 124 nt |
| IRs (inverted repeats) | 101..106, 119..124 (TTTAAT..ATTAAA) 91..99, 113..121 (AATAATGTA..TACATTATT) 90..95, 107..112 (AAATAA..TTATTT) 88..93, 105..110 (ATAAAT..ATTTAT) 41..48, 61..68 (AAAAACAA..TTGTTTTT) 39..46, 49..56 (GCAAAAAC..GTTTTTGC) 3..10, 15..22 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 124 nt
GGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTGTTTTTTGAATCATTAGCTTATGTTATAAATAATGTATTTTAATTTATTTTACATTATTAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file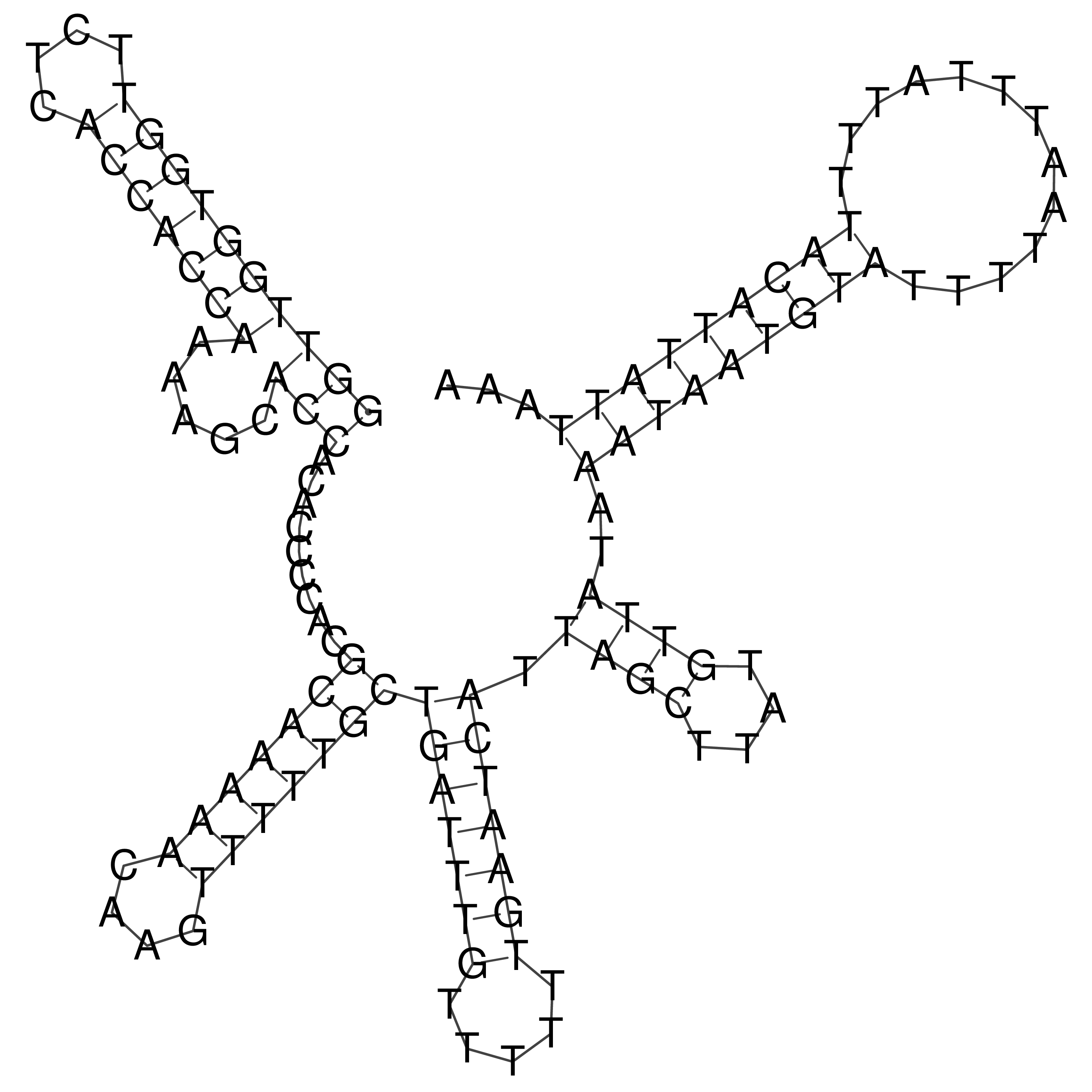
Relaxase
| ID | 9484 | GenBank | WP_256876176 |
| Name | traI_NIZ20_RS24470_231_1_NP_B|plas2 |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 191989.88 Da Isoelectric Point: 5.9597
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTHLLEGRLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRDAADQRAETRTQTPGPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLQEGMAFTPGNTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGRPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSNSLTLRDAQCETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDAMTVVVPGRAEPASLPVSDSPFMALKL
ENGWVETPGHSVSDSAKVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETLLETAISLQKTGLHTPAQQAIHLALPVVESKNLAFSMVDLLTEAKSFAAEGTSFTELGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLLQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRLEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALALVDNVYHRIAGISRDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDTIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKLMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVF
EPKPDREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTEQAVRDIAGLERDRSAISEREAALPESVLREPQRVREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 5563 | GenBank | WP_000124979 |
| Name | WP_000124979_231_1_NP_B|plas2 |
UniProt ID | Q5JBL6 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14456.50 Da Isoelectric Point: 5.2973
MARVNLYISNEVHEKINMIVEKRRQEGARDKDISLSGTASMLLELGLRVYDAQMERKESAFNQTEFNKLL
LECAVKTQSTVAKILGIESLSPHVSGNPKFEYASMVDDIREKVSVEMDRFFPKNDGE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q5JBL6 |
| ID | 5564 | GenBank | WP_001254385 |
| Name | WP_001254385_231_1_NP_B|plas2 |
UniProt ID | A0A1Q9L2K4 |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 8975.08 Da Isoelectric Point: 10.1422
MRRRNARGGISRTVSVYLDEDTNNRLIKAKDRSGRSKTIEVQIRLRDHLKRFPDFYNEEIFREVAEESES
TFKEL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A1Q9L2K4 |
T4CP
| ID | 11038 | GenBank | WP_001064248 |
| Name | traC_NIZ20_RS24375_231_1_NP_B|plas2 |
UniProt ID | _ |
| Length | 875 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 875 a.a. Molecular weight: 99269.03 Da Isoelectric Point: 6.1932
MNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAIP
INGANESIVEALDHMLRTKLPRGIPLCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMKAAA
TQFPLPEGMNLPLTLRHYRVFISYCSPSKKKSRADILEMENLVKIIRASLQGASITTQTVDAQAFIDIVG
EMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAFL
WNVADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGE
LRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLSV
MASPNGNLDEVHEGLLLQAVRASWLAKKNRARIDDVVDFLKNARDNDQYVESPTIRSRLDEMIVLLDQYT
ANGTYGQYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGWR
LLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLYPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEGL
SIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 11039 | GenBank | WP_153449148 |
| Name | traD_NIZ20_RS24465_231_1_NP_B|plas2 |
UniProt ID | _ |
| Length | 738 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 738 a.a. Molecular weight: 83934.58 Da Isoelectric Point: 5.0504
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIRSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLATVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTDNPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGESFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPDVPEVVSGEDVTQAEQPQQPQQPQQ
PQQPQQPQQPQQPQQPVSPAINDKKSDSGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAAYE
AWQQENHPDIQQQMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 26165..52890
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| NIZ20_RS24255 (NIZ20_24225) | 22130..22564 | + | 435 | WP_000845873 | conjugation system SOS inhibitor PsiB | - |
| NIZ20_RS24260 (NIZ20_24230) | 22561..23323 | + | 763 | Protein_32 | plasmid SOS inhibition protein A | - |
| NIZ20_RS24265 (NIZ20_24235) | 23292..23480 | - | 189 | WP_001299721 | hypothetical protein | - |
| NIZ20_RS24270 (NIZ20_24240) | 23502..23651 | + | 150 | Protein_34 | plasmid maintenance protein Mok | - |
| NIZ20_RS24275 (NIZ20_24245) | 23593..23718 | + | 126 | WP_001372321 | type I toxin-antitoxin system Hok family toxin | - |
| NIZ20_RS24280 (NIZ20_24250) | 23938..24168 | + | 231 | WP_256876178 | hypothetical protein | - |
| NIZ20_RS24285 (NIZ20_24255) | 24166..24339 | - | 174 | Protein_37 | hypothetical protein | - |
| NIZ20_RS24290 (NIZ20_24260) | 24409..24615 | + | 207 | WP_000547971 | hypothetical protein | - |
| NIZ20_RS24295 (NIZ20_24265) | 24640..24927 | + | 288 | WP_000107535 | hypothetical protein | - |
| NIZ20_RS24300 (NIZ20_24270) | 25047..25868 | + | 822 | WP_001234469 | DUF932 domain-containing protein | - |
| NIZ20_RS24305 (NIZ20_24275) | 26165..26812 | - | 648 | WP_256876177 | transglycosylase SLT domain-containing protein | virB1 |
| NIZ20_RS24310 (NIZ20_24280) | 27089..27472 | + | 384 | WP_000124979 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| NIZ20_RS24315 (NIZ20_24285) | 27663..28349 | + | 687 | WP_000332493 | PAS domain-containing protein | - |
| NIZ20_RS24320 (NIZ20_24290) | 28443..28670 | + | 228 | WP_001254385 | conjugal transfer relaxosome protein TraY | - |
| NIZ20_RS24325 (NIZ20_24295) | 28704..29063 | + | 360 | WP_000340272 | type IV conjugative transfer system pilin TraA | - |
| NIZ20_RS24330 (NIZ20_24300) | 29078..29389 | + | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| NIZ20_RS24335 (NIZ20_24305) | 29411..29977 | + | 567 | WP_000399792 | type IV conjugative transfer system protein TraE | traE |
| NIZ20_RS24340 (NIZ20_24310) | 29964..30692 | + | 729 | WP_001230787 | type-F conjugative transfer system secretin TraK | traK |
| NIZ20_RS24345 (NIZ20_24315) | 30692..32119 | + | 1428 | WP_000146685 | F-type conjugal transfer pilus assembly protein TraB | traB |
| NIZ20_RS24350 (NIZ20_24320) | 32109..32681 | + | 573 | WP_000002794 | conjugal transfer pilus-stabilizing protein TraP | - |
| NIZ20_RS24355 (NIZ20_24325) | 32678..32995 | + | 318 | WP_001057292 | conjugal transfer protein TrbD | virb4 |
| NIZ20_RS24360 (NIZ20_24330) | 32992..33243 | + | 252 | WP_001038341 | conjugal transfer protein TrbG | - |
| NIZ20_RS24365 (NIZ20_24335) | 33240..33755 | + | 516 | WP_000809906 | type IV conjugative transfer system lipoprotein TraV | traV |
| NIZ20_RS24370 (NIZ20_24340) | 33889..34110 | + | 222 | WP_001278689 | conjugal transfer protein TraR | - |
| NIZ20_RS24375 (NIZ20_24345) | 34270..36897 | + | 2628 | WP_001064248 | type IV secretion system protein TraC | virb4 |
| NIZ20_RS24380 (NIZ20_24350) | 36894..37280 | + | 387 | WP_000214096 | type-F conjugative transfer system protein TrbI | - |
| NIZ20_RS24385 (NIZ20_24355) | 37277..37909 | + | 633 | WP_001203745 | type-F conjugative transfer system protein TraW | traW |
| NIZ20_RS24390 (NIZ20_24360) | 37906..38898 | + | 993 | WP_000830839 | conjugal transfer pilus assembly protein TraU | traU |
| NIZ20_RS24395 (NIZ20_24365) | 38924..39385 | + | 462 | WP_000277845 | hypothetical protein | - |
| NIZ20_RS24400 (NIZ20_24370) | 39415..39867 | + | 453 | WP_000069427 | hypothetical protein | - |
| NIZ20_RS24405 (NIZ20_24375) | 39895..40533 | + | 639 | WP_001104248 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| NIZ20_RS24410 (NIZ20_24380) | 40530..42338 | + | 1809 | WP_000821818 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| NIZ20_RS24415 (NIZ20_24385) | 42365..42622 | + | 258 | WP_000864325 | conjugal transfer protein TrbE | - |
| NIZ20_RS24420 (NIZ20_24390) | 42615..43358 | + | 744 | WP_001030376 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| NIZ20_RS24425 (NIZ20_24395) | 43374..43721 | + | 348 | WP_001287895 | conjugal transfer protein TrbA | - |
| NIZ20_RS24430 (NIZ20_24400) | 43840..44124 | + | 285 | WP_000624109 | type-F conjugative transfer system pilin chaperone TraQ | - |
| NIZ20_RS24435 (NIZ20_24405) | 44111..44656 | + | 546 | WP_000059847 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| NIZ20_RS24440 (NIZ20_24410) | 44586..44948 | + | 363 | WP_001443292 | P-type conjugative transfer protein TrbJ | - |
| NIZ20_RS24445 (NIZ20_24415) | 44948..46321 | + | 1374 | WP_001137358 | conjugal transfer pilus assembly protein TraH | traH |
| NIZ20_RS24450 (NIZ20_24420) | 46318..49140 | + | 2823 | WP_011251382 | conjugal transfer mating-pair stabilization protein TraG | traG |
| NIZ20_RS24455 (NIZ20_24425) | 49155..49658 | + | 504 | WP_000605510 | hypothetical protein | - |
| NIZ20_RS24460 (NIZ20_24430) | 49690..50421 | + | 732 | WP_000850424 | conjugal transfer complement resistance protein TraT | - |
| NIZ20_RS24465 (NIZ20_24435) | 50674..52890 | + | 2217 | WP_153449148 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 15234 | GenBank | NZ_CP099885 |
| Plasmid name | 231_1_NP_B|plas2 | Incompatibility group | IncFII |
| Plasmid size | 89097 bp | Coordinate of oriT [Strand] | 26737..26860 [+] |
| Host baterium | Escherichia albertii strain 231_1_NP_B |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | espL2 |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |