Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114782 |
| Name | oriT_UNC_SA44|p1 |
| Organism | Staphylococcus aureus strain UNC_SA44 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP132369 (23324..23359 [-], 36 nt) |
| oriT length | 36 nt |
| IRs (inverted repeats) | 4..10, 14..20 (GCGAACG..CGTTCGC) |
| Location of nic site | 29..30 |
| Conserved sequence flanking the nic site |
GTGCGCCCTT |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 36 nt
>oriT_UNC_SA44|p1
CACGCGAACGGAACGTTCGCATAAGTGCGCCCTTAC
CACGCGAACGGAACGTTCGCATAAGTGCGCCCTTAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file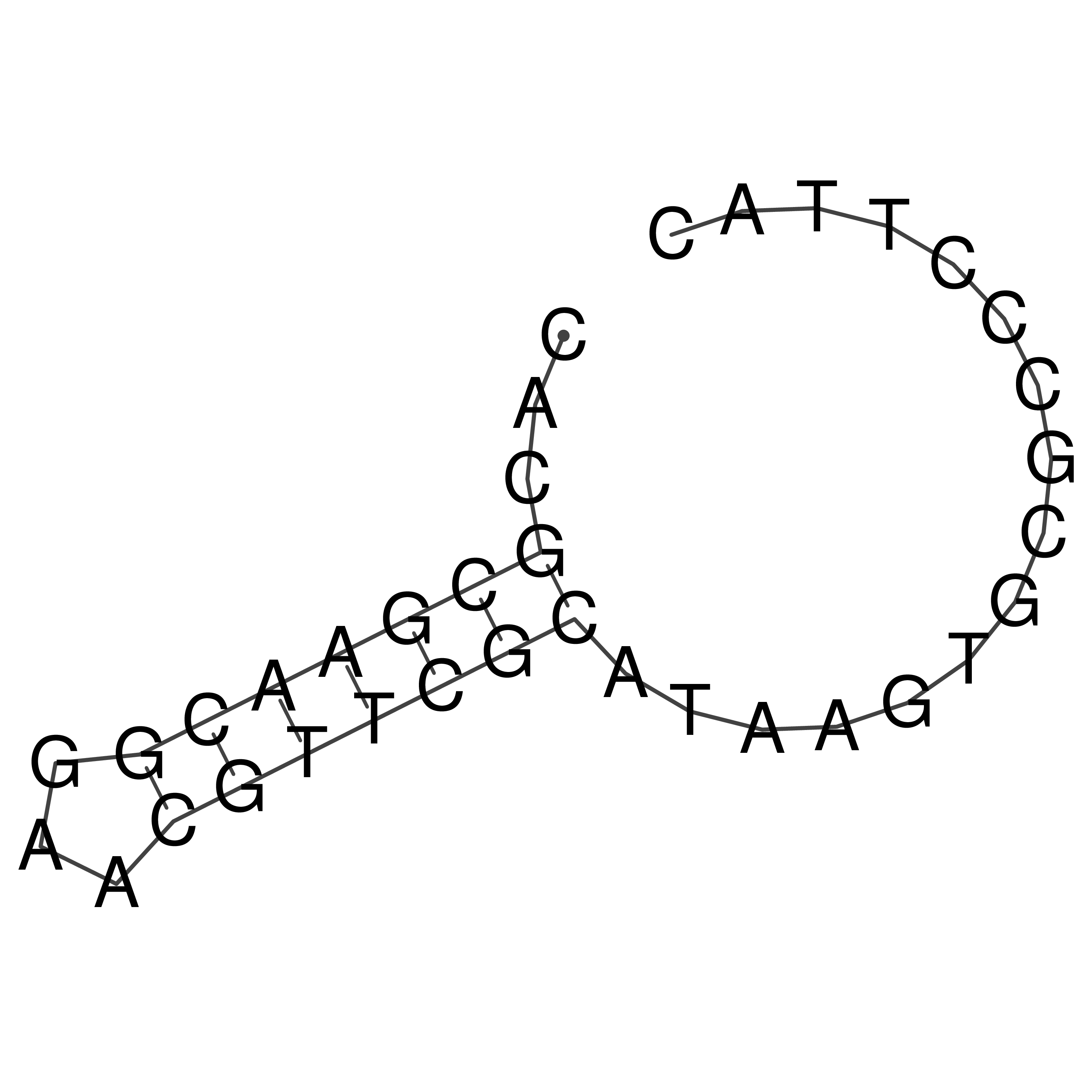
Relaxase
| ID | 9470 | GenBank | WP_306178955 |
| Name | MobA_MobL_RAM31_RS13480_UNC_SA44|p1 |
UniProt ID | _ |
| Length | 665 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 665 a.a. Molecular weight: 78907.88 Da Isoelectric Point: 5.4616
>WP_306178955.1 MobA/MobL family protein [Staphylococcus aureus]
MAMYHFQNKFVSKANGQSATAKSAYNSASRIKDFKENQFKDYSNKQCDYSEILLPNNADDKFKDREYLWN
KVHDVENRKNSQVAREIIIGLPNEFDPNSNIELAKEFAESLSNEGMIVDLNIHKINEENPHAHLLCTLRG
LDKNNEFEPKRKGNDYIRDWNTKEKHNEWRKRWENVQNKHLEKNGFSVRVSADSYKNQNIDLEPTKKEGW
KARKFEDETGKKSSISKHNESIKKRNQQKIDKMFNEVDDMKSHKLNAFSYMNKSDSTTLKNMAKDLKIYV
TPINMYKENERLYDLKQKTSLITDDEDRLNKIEDIEDRQKKLESINEVFEKQAGIFFDKNYPDQSLNYSD
DEKIFITRTILNDRDVLPANNELEDIVKEKRIKEAQISLNTVLGNRDISLESIEKESNFFADKLSNILEK
NNLSFDDVLENKHEGMEDSLKIDYYTNKLEVFRNAENILEDYYDVQIKELFTDDEDYKAFNEVTDIKEKQ
QLIDFKTYHGTENTIEMLETGNFIPKYSDEDRKYITEQVKLLQEKEFKPNKNQHDKFVFGAIQKKLLSEY
DFDYSDNNDLKHLYQESNEVGDEISKDNIEEFYEGNDILVDKETYNNYNRKQQAYGLINAGLDSMIFNFN
EIFRERMPKYINHQYKGKNHSKQRHELKNKRGMHL
MAMYHFQNKFVSKANGQSATAKSAYNSASRIKDFKENQFKDYSNKQCDYSEILLPNNADDKFKDREYLWN
KVHDVENRKNSQVAREIIIGLPNEFDPNSNIELAKEFAESLSNEGMIVDLNIHKINEENPHAHLLCTLRG
LDKNNEFEPKRKGNDYIRDWNTKEKHNEWRKRWENVQNKHLEKNGFSVRVSADSYKNQNIDLEPTKKEGW
KARKFEDETGKKSSISKHNESIKKRNQQKIDKMFNEVDDMKSHKLNAFSYMNKSDSTTLKNMAKDLKIYV
TPINMYKENERLYDLKQKTSLITDDEDRLNKIEDIEDRQKKLESINEVFEKQAGIFFDKNYPDQSLNYSD
DEKIFITRTILNDRDVLPANNELEDIVKEKRIKEAQISLNTVLGNRDISLESIEKESNFFADKLSNILEK
NNLSFDDVLENKHEGMEDSLKIDYYTNKLEVFRNAENILEDYYDVQIKELFTDDEDYKAFNEVTDIKEKQ
QLIDFKTYHGTENTIEMLETGNFIPKYSDEDRKYITEQVKLLQEKEFKPNKNQHDKFVFGAIQKKLLSEY
DFDYSDNNDLKHLYQESNEVGDEISKDNIEEFYEGNDILVDKETYNNYNRKQQAYGLINAGLDSMIFNFN
EIFRERMPKYINHQYKGKNHSKQRHELKNKRGMHL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 11016 | GenBank | WP_000209436 |
| Name | t4cp2_RAM31_RS13370_UNC_SA44|p1 |
UniProt ID | _ |
| Length | 546 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 546 a.a. Molecular weight: 62672.60 Da Isoelectric Point: 7.6569
>WP_000209436.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Bacilli]
MTTLLGELESKATSINKKNLVIQDFDTKCPNRNIFVVGGPGSYKSAGYVIPNVIVKNQQSIVVTDPSGEV
YEKTSNIKRMQGFDVRVVNFKNFLASDRQNFFDYIDKDSDCSIVAELIIKSAGDSKINKDVWYKASVGLL
NSLLLYAKYEFEPEKRTIGNIIKFIQNNKPNQDEEGSVELDKRFNELSKDHPARESYEFGFAVSEGRTRA
SIILTFVADLRNYIDNEIRSYTSDSDFDLRDVGLRKTIIYVMLPVLGNTVQSLSSLFFSQMFQQLYRLGD
ENGARLPVPVDFLLDEWPNIGAIPDYEETLATCRKYGISITTIVQSISQAVDKYNKDKANAIIGNHAVIL
CLGNVNNDTAKYIKEELGNATVEFETSSEGSSSGKDSRSKSSNKNVSYTGRALLNEDEISNMQQDKAILI
TKNRNPKIIKKLAYFKMFPNLTETYIAPQNEYKRKKNQSMIDKHNRLREEWDKKQEKNKQQKLEEYKEEQ
RQKELEKEQQPQEEQQNEKVKESKDKNDQEDKKDEQQKINDSKKAFYEKLKQKQAK
MTTLLGELESKATSINKKNLVIQDFDTKCPNRNIFVVGGPGSYKSAGYVIPNVIVKNQQSIVVTDPSGEV
YEKTSNIKRMQGFDVRVVNFKNFLASDRQNFFDYIDKDSDCSIVAELIIKSAGDSKINKDVWYKASVGLL
NSLLLYAKYEFEPEKRTIGNIIKFIQNNKPNQDEEGSVELDKRFNELSKDHPARESYEFGFAVSEGRTRA
SIILTFVADLRNYIDNEIRSYTSDSDFDLRDVGLRKTIIYVMLPVLGNTVQSLSSLFFSQMFQQLYRLGD
ENGARLPVPVDFLLDEWPNIGAIPDYEETLATCRKYGISITTIVQSISQAVDKYNKDKANAIIGNHAVIL
CLGNVNNDTAKYIKEELGNATVEFETSSEGSSSGKDSRSKSSNKNVSYTGRALLNEDEISNMQQDKAILI
TKNRNPKIIKKLAYFKMFPNLTETYIAPQNEYKRKKNQSMIDKHNRLREEWDKKQEKNKQQKLEEYKEEQ
RQKELEKEQQPQEEQQNEKVKESKDKNDQEDKKDEQQKINDSKKAFYEKLKQKQAK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 3415..14635
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| RAM31_RS13335 (RAM31_13330) | 1186..1578 | - | 393 | WP_000393259 | GNAT family N-acetyltransferase | - |
| RAM31_RS13340 (RAM31_13335) | 1635..2009 | - | 375 | Protein_2 | IS256 family transposase | - |
| RAM31_RS13345 (RAM31_13340) | 2070..2744 | - | 675 | WP_011117680 | IS6-like element IS257 family transposase | - |
| RAM31_RS13350 (RAM31_13345) | 2782..2949 | - | 168 | WP_000869301 | hypothetical protein | - |
| RAM31_RS13355 (RAM31_13350) | 3006..3398 | - | 393 | WP_000608970 | single-stranded DNA-binding protein | - |
| RAM31_RS13360 (RAM31_13355) | 3415..3900 | - | 486 | WP_258390337 | conjugal transfer protein TrbL family protein | trsL |
| RAM31_RS13365 (RAM31_13360) | 3855..4331 | - | 477 | WP_258390336 | hypothetical protein | trsL |
| RAM31_RS13370 (RAM31_13365) | 4409..6049 | - | 1641 | WP_000209436 | VirD4-like conjugal transfer protein, CD1115 family | - |
| RAM31_RS13375 (RAM31_13370) | 6046..6510 | - | 465 | WP_000715273 | hypothetical protein | trsJ |
| RAM31_RS13380 (RAM31_13375) | 6526..8628 | - | 2103 | WP_001094109 | DNA topoisomerase III | - |
| RAM31_RS13385 (RAM31_13380) | 8644..9129 | - | 486 | WP_000735569 | TrsH/TraH family protein | - |
| RAM31_RS13390 (RAM31_13385) | 9139..10215 | - | 1077 | WP_000269385 | CHAP domain-containing protein | trsG |
| RAM31_RS13395 (RAM31_13390) | 10233..11513 | - | 1281 | WP_000702361 | membrane protein | trsF |
| RAM31_RS13400 (RAM31_13395) | 11525..13537 | - | 2013 | WP_000874919 | VirB4 family type IV secretion system protein | virb4 |
| RAM31_RS13405 (RAM31_13400) | 13558..14241 | - | 684 | WP_306178959 | TrsD/TraD family conjugative transfer protein | trsD |
| RAM31_RS13410 (RAM31_13405) | 14228..14635 | - | 408 | WP_306178960 | hypothetical protein | trsC |
| RAM31_RS13415 (RAM31_13410) | 14695..15012 | - | 318 | WP_000591622 | CagC family type IV secretion system protein | - |
| RAM31_RS13420 (RAM31_13415) | 15029..16003 | - | 975 | WP_000368849 | conjugative transfer protein TrsA | - |
| RAM31_RS13425 (RAM31_13420) | 16175..16363 | + | 189 | WP_001063224 | hypothetical protein | - |
| RAM31_RS13430 (RAM31_13425) | 16472..17146 | - | 675 | WP_001106019 | IS6-like element IS257 family transposase | - |
| RAM31_RS13435 (RAM31_13430) | 17190..17336 | + | 147 | WP_001817822 | hypothetical protein | - |
| RAM31_RS13440 (RAM31_13435) | 17496..18266 | + | 771 | WP_001795128 | aminoglycoside O-nucleotidyltransferase ANT(4')-Ia | - |
| RAM31_RS13445 (RAM31_13440) | 18593..19267 | - | 675 | WP_001106022 | IS6-like element IS257 family transposase | - |
Host bacterium
| ID | 15217 | GenBank | NZ_CP132369 |
| Plasmid name | UNC_SA44|p1 | Incompatibility group | - |
| Plasmid size | 36305 bp | Coordinate of oriT [Strand] | 23324..23359 [-] |
| Host baterium | Staphylococcus aureus strain UNC_SA44 |
Cargo genes
| Drug resistance gene | aadD |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |