Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114687 |
| Name | oriT_pSR8 |
| Organism | Aminobacter sp. SR38 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP062120 (147988..148086 [-], 99 nt) |
| oriT length | 99 nt |
| IRs (inverted repeats) | 24..31, 34..41 (TAACCGGC..GCCGGTTA) |
| Location of nic site | 60..61 |
| Conserved sequence flanking the nic site |
TCCTGCCCGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 99 nt
GAATAAGGGACAGTGAAGATAGATAACCGGCTCGCCGGTTAGCTAACTTCACACATCCTGCCCGCCTTACGGCGTTAATAACACCAAGGAAAGTCTACA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file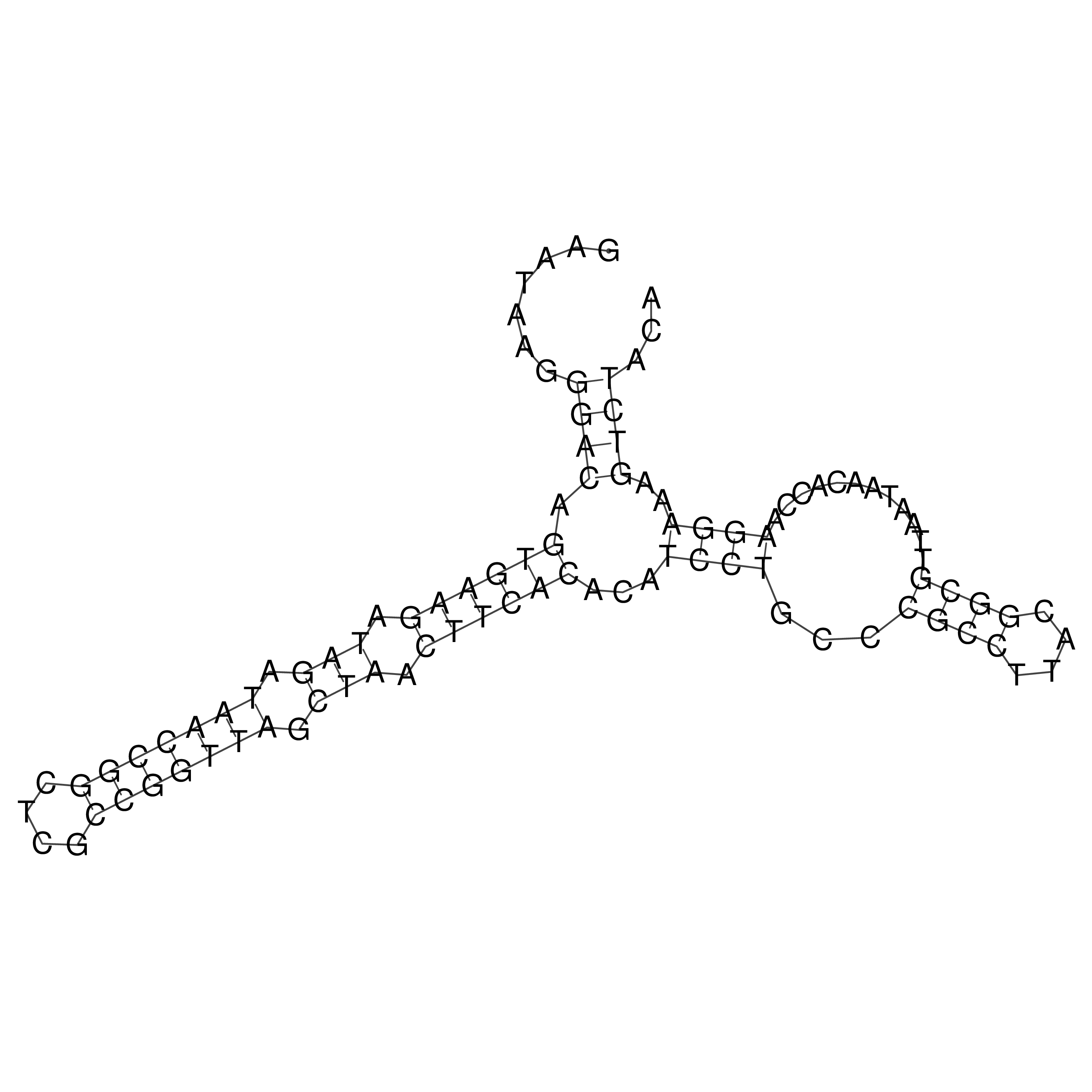
Relaxase
| ID | 9422 | GenBank | WP_011117146 |
| Name | traI_IG197_RS35820_pSR8 |
UniProt ID | Q937B9 |
| Length | 746 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 746 a.a. Molecular weight: 82120.82 Da Isoelectric Point: 10.8213
MIAKHVPMRSLGKSDFAGLANYITDAQSKDHRLGHVQATNCEAGSIQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICVGLGYGEHQRISAVHNDTDNLHIHIAINKIHPTRHTMHEPYYPHRA
LAELCTALERDYGLERDNHEPRKRGAEGRAADMERHAGVESLVGWVKRECLDEIKGAQSWQELHQVMRDN
GMELRVRANGLVFEAGDGTMVKASTVARDLSKPSLEARLGPFEASPERQAQTTAKRQYRKDPIRLRVNTV
ELYAKYKAEQQNLTTARAQALERARHRKDRLIEAAKRSNRLRRATIKVVGEGRANKKLLYAQASKALRSE
IQAINKQFQQERTALYAEHRRRTWADWLKKEAQHGGADALAALRAREAAQGLKGNTIRGEGQAKPGHAPA
VDNITKKGTIIFRAGMSAVRDDGDRLQVSREATREGLQEALRLAMQRYGNRITVNGTVEFKAQMIRAAVD
SQLPITFTDPALESRRQALLNKENTHERTERPEHRGRTGRGAGGPGQRPAADQHATGAAAVARAGDGRPA
AGRGDRADAGLHAATVHRKPDVGRLGRKPPPQSQHRLRALSELGVVRIAGGSEVLLPRDVPRHVEQQGTQ
PDHALRRGISRPGTGVGQTPPGVAAADKYIAEREAKRLKGFDIPKHSRYTAGDGALTFQGTRTIEGQALA
LLKRGDEVMVMPIDQATARRLTRIAVGDAVSITAKGSIKTSKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q937B9 |
T4CP
| ID | 10949 | GenBank | WP_000694765 |
| Name | t4cp2_IG197_RS35150_pSR8 |
UniProt ID | _ |
| Length | 637 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 637 a.a. Molecular weight: 69897.09 Da Isoelectric Point: 8.2993
MKIKMNNAVGPQVRTAKPKPSKLLPVLGAASMVGGLQAATQFFAHTFAYHATLGPNVGHVYAPWSILHWT
YKWYSQYPDEIMKAGSMGMLVSTVGLLGVAVAKVVTSNSSKANEYLHGSARWAEKKDIQAAGLLPRERNV
LEIVTGKAAPTATGVYVGGWQDKDGNFFYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASTSGGVCWNPLDEIRLGTEYEVGDVQNLATLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKAKDDGGTATLPSVDAMLADPNRDIGELWMEMATYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLALYRDPVVARNVSRSDFRIKQLMHEDDPVSLYIVTQPNDKARLRPLVRVMVN
MIVRLLADKMDFEGGRPVAHYKHRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLKSRET
GYGHDESITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTYQEVQRPLLT
PDECLRMPGPKKNAQGEIEEAGDMVIYVAGYPAIYGKQPLYFKDPVFSARAAIPAPKVSDRLRAVAQAET
EGEGITI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 10950 | GenBank | WP_011114047 |
| Name | t4cp2_IG197_RS35465_pSR8 |
UniProt ID | _ |
| Length | 852 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 852 a.a. Molecular weight: 93980.15 Da Isoelectric Point: 6.3901
MIEAIAIAIAVLGAVLLLILFARIRAVDAELKLKKHRAKDAGLADLLNYAAVVDDGVIVGKNGSFMAAWL
YKGDDNASSTEEQREMVSFRINQALAGLGNGWMVHVDAVRRPAPNYSERGVSSFPDSVSFAIDEERRRLF
EGLGAMFEGYFVLTVTWFPPVLAQRKFVELMFDDDAVAPDHTARTTGLIAQFKREITSLESRLSSAVKMT
RLRGQKVVSEEGREVTHDDFLSWLQFCVTGLHHPVMLPSNPMYLDAVIGGQELWGGVVPKIGRKFIQVVA
IEGFPLESTPGVLSALAELPSEYRWSSRFIFMDQHESLKHLDKFRKKWKQKIRGFFDQVFNTNTGSINQD
AAAMVGDAEAAIAEVNSGLVAAGYYTSVVVLMDEDRERLAASALLVEKAVNRLAFAARIETINTLDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSSIWTGSATAPCPMYPPLSPALMHCVTVGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGIIAAQLRRYAGMSIYAFDKGMSMYPLAAGIRAATKGKSGLHFTVAADDDRLAFC
PLQFLETKGDRAWAMEWIDTILALNGVNTTPAQRNEIGNAIMSMHASGARTLSEFSVTIQDEAIREAIKQ
YTVDGSMGHLLDAEEDGLSLSDFTVFEIEELMNLGEKFALPVLLYLFRRIERALKGQPSVIILDEAWLML
GHPAFRAKIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTSALYRRMG
LNARQIEILATAIPKRQYYYVSENGRRLYDLALGPMALAFVGASDKESVATIKSLEAKFGHEWVHEWLAG
RGLNLNDYLEAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 10951 | GenBank | WP_000694765 |
| Name | t4cp2_IG197_RS35825_pSR8 |
UniProt ID | _ |
| Length | 637 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 637 a.a. Molecular weight: 69897.09 Da Isoelectric Point: 8.2993
MKIKMNNAVGPQVRTAKPKPSKLLPVLGAASMVGGLQAATQFFAHTFAYHATLGPNVGHVYAPWSILHWT
YKWYSQYPDEIMKAGSMGMLVSTVGLLGVAVAKVVTSNSSKANEYLHGSARWAEKKDIQAAGLLPRERNV
LEIVTGKAAPTATGVYVGGWQDKDGNFFYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASTSGGVCWNPLDEIRLGTEYEVGDVQNLATLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKAKDDGGTATLPSVDAMLADPNRDIGELWMEMATYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLALYRDPVVARNVSRSDFRIKQLMHEDDPVSLYIVTQPNDKARLRPLVRVMVN
MIVRLLADKMDFEGGRPVAHYKHRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLKSRET
GYGHDESITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTYQEVQRPLLT
PDECLRMPGPKKNAQGEIEEAGDMVIYVAGYPAIYGKQPLYFKDPVFSARAAIPAPKVSDRLRAVAQAET
EGEGITI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 78779..89455
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| IG197_RS35405 (IG197_35405) | 74147..75693 | - | 1547 | Protein_80 | IS91-like element ISPps1 family transposase | - |
| IG197_RS35410 (IG197_35410) | 75802..77007 | - | 1206 | Protein_81 | NAD(P)/FAD-dependent oxidoreductase | - |
| IG197_RS35415 (IG197_35415) | 77153..77482 | + | 330 | Protein_82 | DUF4158 domain-containing protein | - |
| IG197_RS35420 (IG197_35420) | 77525..78160 | - | 636 | WP_011114052 | transglycosylase SLT domain-containing protein | - |
| IG197_RS35425 (IG197_35425) | 78174..78761 | - | 588 | WP_010890138 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| IG197_RS35430 (IG197_35430) | 78779..80734 | - | 1956 | WP_246765562 | P-type conjugative transfer protein TrbL | virB6 |
| IG197_RS35440 (IG197_35440) | 80744..81508 | - | 765 | WP_010890135 | P-type conjugative transfer protein TrbJ | virB5 |
| IG197_RS35445 (IG197_35445) | 81529..82950 | - | 1422 | WP_011114050 | TrbI/VirB10 family protein | virB10 |
| IG197_RS35450 (IG197_35450) | 82955..83443 | - | 489 | WP_006122479 | conjugal transfer protein TrbH | - |
| IG197_RS35455 (IG197_35455) | 83446..84345 | - | 900 | WP_006122480 | P-type conjugative transfer protein TrbG | virB9 |
| IG197_RS35460 (IG197_35460) | 84363..85145 | - | 783 | WP_011114048 | conjugal transfer protein TrbF | virB8 |
| IG197_RS35465 (IG197_35465) | 85142..87700 | - | 2559 | WP_011114047 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| IG197_RS35470 (IG197_35470) | 87697..88008 | - | 312 | WP_001207961 | conjugal transfer protein TrbD | virB3 |
| IG197_RS35475 (IG197_35475) | 88012..88476 | - | 465 | WP_011114046 | conjugal transfer system pilin TrbC | virB2 |
| IG197_RS35480 (IG197_35480) | 88493..89455 | - | 963 | WP_000132020 | P-type conjugative transfer ATPase TrbB | virB11 |
| IG197_RS35485 (IG197_35485) | 89765..90127 | - | 363 | WP_011114045 | transcriptional regulator | - |
| IG197_RS35490 (IG197_35490) | 90241..90582 | + | 342 | WP_000019167 | single-stranded DNA-binding protein | - |
| IG197_RS35495 (IG197_35495) | 90629..91849 | + | 1221 | WP_011114072 | plasmid replication initiator TrfA | - |
| IG197_RS35500 (IG197_35500) | 91962..92396 | - | 435 | WP_003131969 | mercury resistance transcriptional regulator MerR | - |
| IG197_RS35505 (IG197_35505) | 92468..92818 | + | 351 | WP_003131974 | mercuric ion transporter MerT | - |
| IG197_RS35510 (IG197_35510) | 92831..93106 | + | 276 | WP_003131987 | mercury resistance system periplasmic binding protein MerP | - |
Host bacterium
| ID | 15122 | GenBank | NZ_CP062120 |
| Plasmid name | pSR8 | Incompatibility group | IncP1 |
| Plasmid size | 197271 bp | Coordinate of oriT [Strand] | 147988..148086 [-] |
| Host baterium | Aminobacter sp. SR38 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | merE, merD, merB, merR2, merA, merP, merT, merR1, merR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIF24 |