Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114676 |
| Name | oriT_pA |
| Organism | Agrobacterium sp. CGMCC 11546 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP047389 (122892..122923 [-], 32 nt) |
| oriT length | 32 nt |
| IRs (inverted repeats) | 18..23, 27..32 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 32 nt
>oriT_pA
CCAAGGGCGCAATTATACGTCGCGATGCGACG
CCAAGGGCGCAATTATACGTCGCGATGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file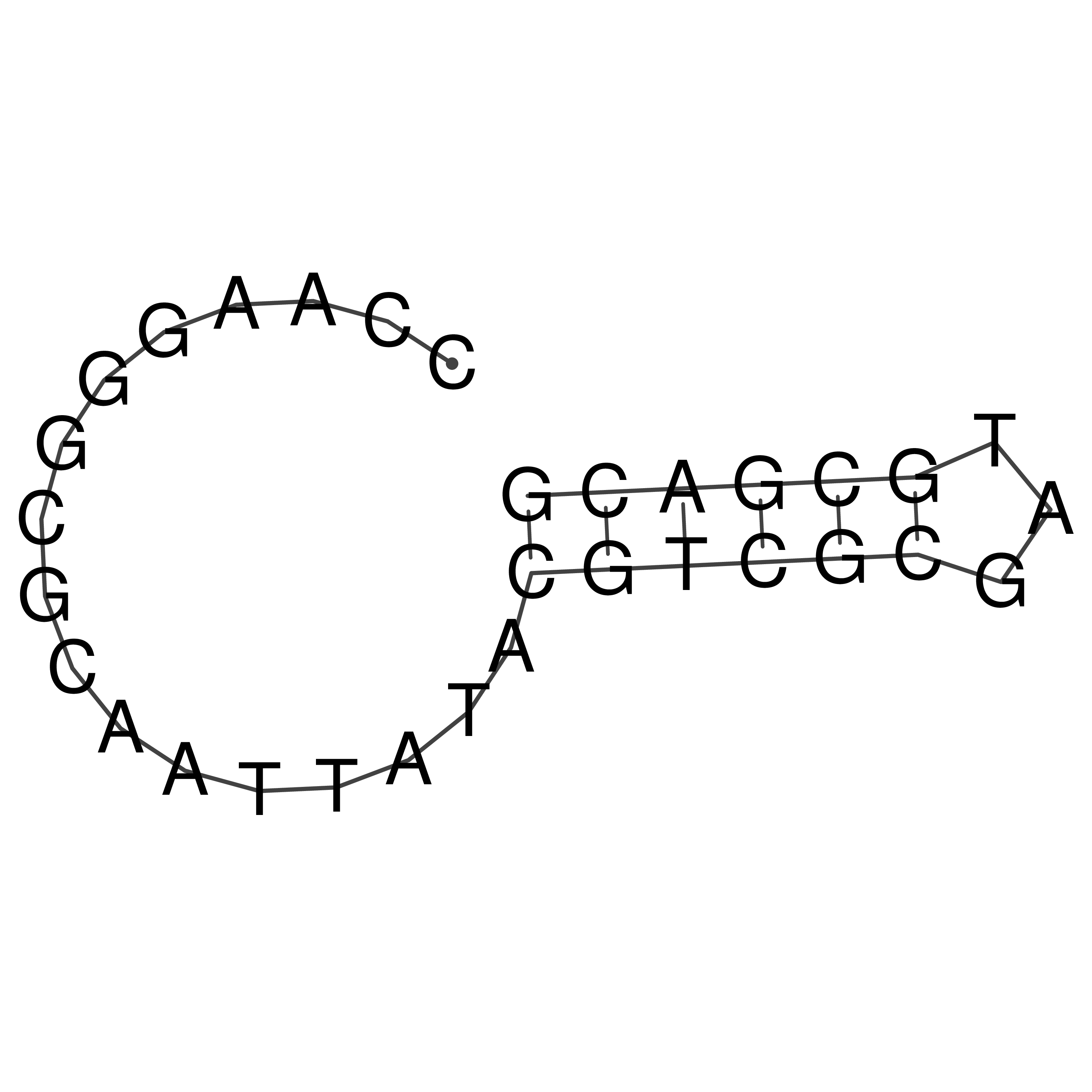
Relaxase
| ID | 9415 | GenBank | WP_006315302 |
| Name | traA_GSF67_RS23905_pA |
UniProt ID | A0AA87AZE6 |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 123738.41 Da Isoelectric Point: 9.7851
>WP_006315302.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [unclassified Agrobacterium]
MAIAHFSASIVSRGSGRSVVLSAAYRHCAKMEFEREARTIDYTRKQRLLHEEFALPADAPKWVRSLIADR
SVSGASEAFWNKVEAFEKRADAQLARDLTIALPLELSADQNIALVRDFVEKHILAKGMVADWVFHDNPGN
PHIHLMTTLRPLSEDGFGSKKIAVMGDDGQPVRTKSGKILYELWAGSTEDFNVLRDGWFERQNHHLALGG
IDLKIDGRSYEKQGIDLEPTIHLGVGTKAIERKAEQQGVRPELERIELNDARRAENARRILKDPAIVLDL
IMREKSVFNERDVAKVLHRYVDDPAVFQQLMLRIMLNPEVLRLQRDTIDFATGEMVPARYSTQAMIRLEA
TMARQATWLSEREGRAVSGTILEGTFQRHEQLSEEQRTAIERIAGPARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIESRTLASWELRWNRGRDVLDDKTIFVMDEAGMVASKQMAGFVD
AVVRAGAKIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQRADWMRKASLDLARGNVEKALITYQ
REGRVLGSRLKSEAVEYLIADWNRDYDQTKTTLILAHLRRDVRMLNVMAREKLIERGVVGEGHVFKTADG
VRRFDAGDQIVFLKNEGSLGVKNGMIAHIAEAQPNRIVAVVGEGDHRRHVVVEQRFYNNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGTRSFSFNGGLAKVLSRRQAKETTLDYERGQLY
REALRFSENRGLYIVQVARTLVRDRLDWTLREKAKLVELGQRLAAFAARLGITQSPNPHTMKEAAPMVAG
TKTFAGSVADTVGDKLGADPALKRQWEEVSARFAYVFADPETAFRAMNFDAVLAEKEMAKQVLRKIEVEP
ASIGPLKGKTGILATRSAREARRVAEVNIPALKRDLEQYLRMREMVTQRLQADEQALRQRVSIDIPALSP
AAHVVLERVRDAIDRNDLPSAMAYALSNRETKLEIDEFNKAVTERFGERTLLSNAAHEPSGKLYEELSKG
MKPEQKEELKQAWPVMRTAQQLSAHERTVQSLRQAEEQRLTQRQTPVRKQ
MAIAHFSASIVSRGSGRSVVLSAAYRHCAKMEFEREARTIDYTRKQRLLHEEFALPADAPKWVRSLIADR
SVSGASEAFWNKVEAFEKRADAQLARDLTIALPLELSADQNIALVRDFVEKHILAKGMVADWVFHDNPGN
PHIHLMTTLRPLSEDGFGSKKIAVMGDDGQPVRTKSGKILYELWAGSTEDFNVLRDGWFERQNHHLALGG
IDLKIDGRSYEKQGIDLEPTIHLGVGTKAIERKAEQQGVRPELERIELNDARRAENARRILKDPAIVLDL
IMREKSVFNERDVAKVLHRYVDDPAVFQQLMLRIMLNPEVLRLQRDTIDFATGEMVPARYSTQAMIRLEA
TMARQATWLSEREGRAVSGTILEGTFQRHEQLSEEQRTAIERIAGPARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIESRTLASWELRWNRGRDVLDDKTIFVMDEAGMVASKQMAGFVD
AVVRAGAKIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQRADWMRKASLDLARGNVEKALITYQ
REGRVLGSRLKSEAVEYLIADWNRDYDQTKTTLILAHLRRDVRMLNVMAREKLIERGVVGEGHVFKTADG
VRRFDAGDQIVFLKNEGSLGVKNGMIAHIAEAQPNRIVAVVGEGDHRRHVVVEQRFYNNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGTRSFSFNGGLAKVLSRRQAKETTLDYERGQLY
REALRFSENRGLYIVQVARTLVRDRLDWTLREKAKLVELGQRLAAFAARLGITQSPNPHTMKEAAPMVAG
TKTFAGSVADTVGDKLGADPALKRQWEEVSARFAYVFADPETAFRAMNFDAVLAEKEMAKQVLRKIEVEP
ASIGPLKGKTGILATRSAREARRVAEVNIPALKRDLEQYLRMREMVTQRLQADEQALRQRVSIDIPALSP
AAHVVLERVRDAIDRNDLPSAMAYALSNRETKLEIDEFNKAVTERFGERTLLSNAAHEPSGKLYEELSKG
MKPEQKEELKQAWPVMRTAQQLSAHERTVQSLRQAEEQRLTQRQTPVRKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 10939 | GenBank | WP_006315299 |
| Name | traG_GSF67_RS23920_pA |
UniProt ID | _ |
| Length | 656 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 656 a.a. Molecular weight: 70090.21 Da Isoelectric Point: 9.7397
>WP_006315299.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [unclassified Agrobacterium]
MTVNRLLLGVLPAIIMLVVMFGTTGMEHRLAALGTSAQVKLFLGRAGLGLPYVAGAAIGVISLFAANGSA
NIKTAALSVLAGNCAVIVIAVVREAIRLAGIASSVPVGQSVLGYADPATMVGASAAFFSAVFALRVAIKG
NAAFAQGGPTRVGGKRAVHGDANWMSLAKAAKLFPDAGGIVVGERYRVDHDRVAATAFRADDLQSWGGGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSSEVAPMVVDYRRKAGRKVIVLD
PANPATGFNALDWIGRFGGTKEEDIVAVATWIMTDNARSASARDDFFRASAMQLLTALIADVCLSGHTGA
KDQTLRRVRVNLSEPEPKLRERLTRIYEQSESTFVRENVAVFVNMTPETFSGVYANAVKETHWLSYTNYA
ALVSGDSFSTDELATGVTDIFIALDLKVLEAHPGLARVVIGSFLNAIYNRNGEVAGRTLFLLDEVARLGY
LRILETARDAGRKYGISLALIFQSIGQMREAYGGRDASSKWFESASWISFAAINDPETAEYISKRCGDTT
VEVDQTNRSSGMKGSSRSRSKQLTRRPLILPHEVLRMRGDEQIVFTAGNPPLRCGRAIWFRRKDMSACVG
GNRFHKQATDGVKSYKAAPNTDNEET
MTVNRLLLGVLPAIIMLVVMFGTTGMEHRLAALGTSAQVKLFLGRAGLGLPYVAGAAIGVISLFAANGSA
NIKTAALSVLAGNCAVIVIAVVREAIRLAGIASSVPVGQSVLGYADPATMVGASAAFFSAVFALRVAIKG
NAAFAQGGPTRVGGKRAVHGDANWMSLAKAAKLFPDAGGIVVGERYRVDHDRVAATAFRADDLQSWGGGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSSEVAPMVVDYRRKAGRKVIVLD
PANPATGFNALDWIGRFGGTKEEDIVAVATWIMTDNARSASARDDFFRASAMQLLTALIADVCLSGHTGA
KDQTLRRVRVNLSEPEPKLRERLTRIYEQSESTFVRENVAVFVNMTPETFSGVYANAVKETHWLSYTNYA
ALVSGDSFSTDELATGVTDIFIALDLKVLEAHPGLARVVIGSFLNAIYNRNGEVAGRTLFLLDEVARLGY
LRILETARDAGRKYGISLALIFQSIGQMREAYGGRDASSKWFESASWISFAAINDPETAEYISKRCGDTT
VEVDQTNRSSGMKGSSRSRSKQLTRRPLILPHEVLRMRGDEQIVFTAGNPPLRCGRAIWFRRKDMSACVG
GNRFHKQATDGVKSYKAAPNTDNEET
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 15111 | GenBank | NZ_CP047389 |
| Plasmid name | pA | Incompatibility group | - |
| Plasmid size | 302198 bp | Coordinate of oriT [Strand] | 122892..122923 [-] |
| Host baterium | Agrobacterium sp. CGMCC 11546 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | PFL_4714 |
| Symbiosis gene | - |
| Anti-CRISPR | - |