Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114655 |
| Name | oriT_MGH 3|unnamed1 |
| Organism | Enterobacter sp. MGH 3 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP072954 (186095..186197 [-], 103 nt) |
| oriT length | 103 nt |
| IRs (inverted repeats) | 89..94, 96..101 (AAATAA..TTATTT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 103 nt
>oriT_MGH 3|unnamed1
CTCACCACCAAAAGCTCCACACCCCACGCAAAATTAAAGTTTTTCTGATTGGGTATTCAAATCATGCAGTTAGTGAAAAAGTAATGTGAAATAATTTATTTTA
CTCACCACCAAAAGCTCCACACCCCACGCAAAATTAAAGTTTTTCTGATTGGGTATTCAAATCATGCAGTTAGTGAAAAAGTAATGTGAAATAATTTATTTTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file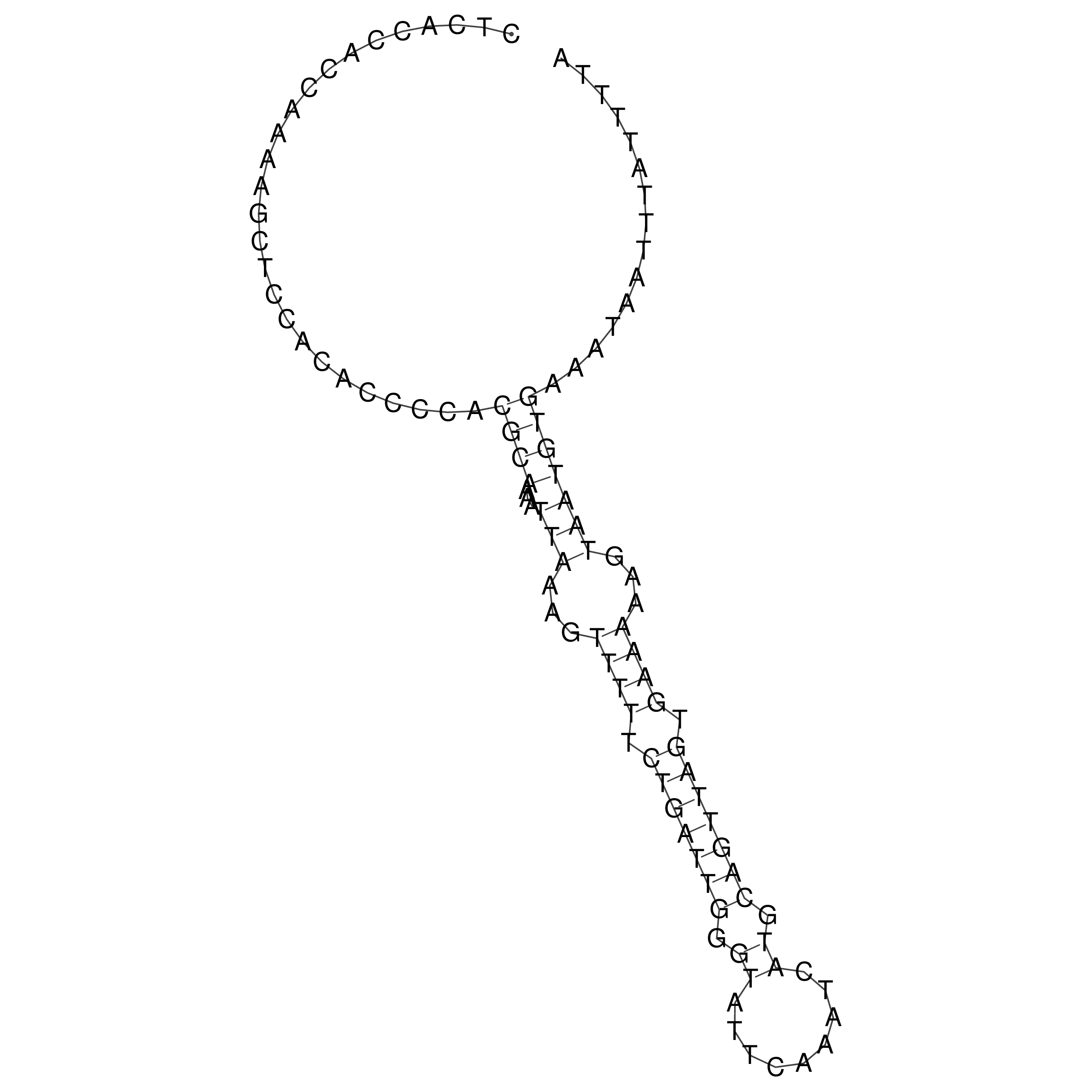
Relaxase
| ID | 9400 | GenBank | WP_022652257 |
| Name | traI_IBG19_RS24800_MGH 3|unnamed1 |
UniProt ID | A0A3S5I3M3 |
| Length | 1746 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1746 a.a. Molecular weight: 189292.49 Da Isoelectric Point: 6.1121
>WP_022652257.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacterales]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGKGAEALGLSGGVDQKVFTKVLEGRLPDGSDLSRTQ
DGANKHRPGYDFTFSAPKSVSVMAMLGGDKRLIEAHNRAVETAIKQIEAMASTRVMTEGRSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANATQHGDEWRTLSSDKVGKTGFIENIYANQIAFGRLYRAALKDDVAA
MGYETETVGKHGMWELKGVPTEPYSSRSRTISEAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMHT
LKETGFDIKAYREAADLRVVQGNIPATTPEAIDINSSVGQAIAMLSDRRARFTYSELLATTLGQLPARSG
MVEMARDGIDAAIKNEQLIPLDKEKGLFTSNIHVLDELSVSAMTRDLQRTGQADIFPDKSVPRSRSYSDA
VSVLAQDRPPVAIISGQGGAAGQRERVAELTMMAREQGRDVQIIAADRRSRSALMQDERLSGEHITDRRG
LTEGMAFTPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTQVSVISEPDRRQRYDRLAADFARSVRAGEHSVAQVTGPREQAVLAGVVREALKAEKVLGEREVSIT
TLEPVWLDSRHQQVRDHYREGMVMERWNAEERSRERFVIDRVTGRNNSLTLRNAQGETRVTRLSELNSSW
SLYRTGTLQVAEGDRLAVLGQAQGTRLRGGDTVTVKALGEAGLVVSLPGRKADVVLPAGDSPFTAAKVVQ
GWVESPGRSVSDSATVFASLSQRELDNTTLNKLALSGRAVRLYSAQTAQKTTEKLSRQSAWSVVSEQIKD
AAGHDRLDDALAHQKAGLHTPEQQAIHLAIPVLEGNGLAFTKPQLMAAAKEFEQDSLSLQAIEKEADRQV
RSGDLLSVPVSPGNGLQLLVSRQSYNAEKSIIRHVLEGKEAVTPLMDKVPDAQLAGLTDGQRNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEREQPRVIGVAPTHRAVSEMRDAGVPETQTLAAFIHDTQ
QQLRGGERTDFSNVLFLVDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DVAVMQEIVRQTPELKPAVYSLISRDIGSALTTIESVVPAQVPRRADAWQPDSSVMEFSREREEAIAKAV
AAGDLMPGGQPATLLEAIVKDYTGRTSEAQAQTIVITALNADRRQVNAMIHDARQGAGEVGEKEVTLPVL
TPANIRDGELRRMATWEASRDSLVLLDSTYYSIEALDGEAHLVMLKDAQGNTRSLSPAQAATEGVTLYRQ
DTITVSEGDRMRFSKSDNERGFVANSVWSVSEIKGDSVTLSDGKQTRTLTPSAEQAEQHIDLAYAVTVEG
SQGASEPFSISLQGTDGGRKRMVSFESAYVALSRMKQHAQVYTDNRDKWVAAMEKSQAKSTAHDILEPRG
DRAVANAARLTATAKALGEVPAGRAALRQAGLQPEGSMAKYISPGRKYPQPHVALPAFDRNGRKAGVWLS
ALTSGDGQLKGLAGEGRVMGSGDAAFAGLQASRNGESLLARDMEEGVRLARENPQSGVVVRLEGEARPWN
PGAITGGRVWADGLPDATGTQPAETVPPEVLAQQAREAEVQRELDKRAEEAVREMARSGDRPAGGADVAV
RDVVRDMDRIKETPASPLTLPDAPEERRRDEAVSQVVRESVQRDRLQQMERETVRDLEREKTLGGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGKGAEALGLSGGVDQKVFTKVLEGRLPDGSDLSRTQ
DGANKHRPGYDFTFSAPKSVSVMAMLGGDKRLIEAHNRAVETAIKQIEAMASTRVMTEGRSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANATQHGDEWRTLSSDKVGKTGFIENIYANQIAFGRLYRAALKDDVAA
MGYETETVGKHGMWELKGVPTEPYSSRSRTISEAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMHT
LKETGFDIKAYREAADLRVVQGNIPATTPEAIDINSSVGQAIAMLSDRRARFTYSELLATTLGQLPARSG
MVEMARDGIDAAIKNEQLIPLDKEKGLFTSNIHVLDELSVSAMTRDLQRTGQADIFPDKSVPRSRSYSDA
VSVLAQDRPPVAIISGQGGAAGQRERVAELTMMAREQGRDVQIIAADRRSRSALMQDERLSGEHITDRRG
LTEGMAFTPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTQVSVISEPDRRQRYDRLAADFARSVRAGEHSVAQVTGPREQAVLAGVVREALKAEKVLGEREVSIT
TLEPVWLDSRHQQVRDHYREGMVMERWNAEERSRERFVIDRVTGRNNSLTLRNAQGETRVTRLSELNSSW
SLYRTGTLQVAEGDRLAVLGQAQGTRLRGGDTVTVKALGEAGLVVSLPGRKADVVLPAGDSPFTAAKVVQ
GWVESPGRSVSDSATVFASLSQRELDNTTLNKLALSGRAVRLYSAQTAQKTTEKLSRQSAWSVVSEQIKD
AAGHDRLDDALAHQKAGLHTPEQQAIHLAIPVLEGNGLAFTKPQLMAAAKEFEQDSLSLQAIEKEADRQV
RSGDLLSVPVSPGNGLQLLVSRQSYNAEKSIIRHVLEGKEAVTPLMDKVPDAQLAGLTDGQRNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEREQPRVIGVAPTHRAVSEMRDAGVPETQTLAAFIHDTQ
QQLRGGERTDFSNVLFLVDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DVAVMQEIVRQTPELKPAVYSLISRDIGSALTTIESVVPAQVPRRADAWQPDSSVMEFSREREEAIAKAV
AAGDLMPGGQPATLLEAIVKDYTGRTSEAQAQTIVITALNADRRQVNAMIHDARQGAGEVGEKEVTLPVL
TPANIRDGELRRMATWEASRDSLVLLDSTYYSIEALDGEAHLVMLKDAQGNTRSLSPAQAATEGVTLYRQ
DTITVSEGDRMRFSKSDNERGFVANSVWSVSEIKGDSVTLSDGKQTRTLTPSAEQAEQHIDLAYAVTVEG
SQGASEPFSISLQGTDGGRKRMVSFESAYVALSRMKQHAQVYTDNRDKWVAAMEKSQAKSTAHDILEPRG
DRAVANAARLTATAKALGEVPAGRAALRQAGLQPEGSMAKYISPGRKYPQPHVALPAFDRNGRKAGVWLS
ALTSGDGQLKGLAGEGRVMGSGDAAFAGLQASRNGESLLARDMEEGVRLARENPQSGVVVRLEGEARPWN
PGAITGGRVWADGLPDATGTQPAETVPPEVLAQQAREAEVQRELDKRAEEAVREMARSGDRPAGGADVAV
RDVVRDMDRIKETPASPLTLPDAPEERRRDEAVSQVVRESVQRDRLQQMERETVRDLEREKTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 10917 | GenBank | WP_022652256 |
| Name | traD_IBG19_RS24805_MGH 3|unnamed1 |
UniProt ID | _ |
| Length | 736 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 736 a.a. Molecular weight: 82566.18 Da Isoelectric Point: 5.1609
>WP_022652256.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacterales]
MSFNAKDMTQGGQIASMRIRMFSQIANIIVYCLVILFGIIVAITLSVKLTWQTFVNGMVYYWCQTLAPMR
DIFKSQPVYTIRYYGHELKYNAQQILQDKYIQWCADQVWTAFILAAIIAAVVCVITFFVTTWILGHQGKQ
QSEDEITGGRQLTDDPKAVAKMLRRDGLASDIKIGELPLIKDSEIQNFCLHGTVGAGKSEVIRRLMNYAR
QRGDMVIVYDRSCEFVKSYYDPATDKILNPLDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLKNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIENNGESFTIRDWMRGVREDGKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDVQTLPDLSCYVTLPGPYPAVKMAL
KYQQRPKVAPEFIPRTIDGKAEERLNAALAAREHESRSIAMLFEPDADVVAPEGGAENVGQPQQPQQPQQ
PQQPQQPQQPQQPQQLTPAPRAADAASVAGAAGAGGVEPELKTKAEEAEQLPPGISESGEVVDMAAWEAW
QAEQDELSPQERQRREEVNINVHRAPEKDLEPGGYI
MSFNAKDMTQGGQIASMRIRMFSQIANIIVYCLVILFGIIVAITLSVKLTWQTFVNGMVYYWCQTLAPMR
DIFKSQPVYTIRYYGHELKYNAQQILQDKYIQWCADQVWTAFILAAIIAAVVCVITFFVTTWILGHQGKQ
QSEDEITGGRQLTDDPKAVAKMLRRDGLASDIKIGELPLIKDSEIQNFCLHGTVGAGKSEVIRRLMNYAR
QRGDMVIVYDRSCEFVKSYYDPATDKILNPLDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLKNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIENNGESFTIRDWMRGVREDGKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDVQTLPDLSCYVTLPGPYPAVKMAL
KYQQRPKVAPEFIPRTIDGKAEERLNAALAAREHESRSIAMLFEPDADVVAPEGGAENVGQPQQPQQPQQ
PQQPQQPQQPQQPQQLTPAPRAADAASVAGAAGAGGVEPELKTKAEEAEQLPPGISESGEVVDMAAWEAW
QAEQDELSPQERQRREEVNINVHRAPEKDLEPGGYI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 10918 | GenBank | WP_008324134 |
| Name | traC_IBG19_RS24880_MGH 3|unnamed1 |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 98841.51 Da Isoelectric Point: 5.7676
>WP_008324134.1 MULTISPECIES: type IV secretion system protein TraC [Enterobacterales]
MSNPLDSVTRTVNSLLTALKMPDASSEANDVLGQMRFPQFSRVLPYRDYDAETGLFMNEKTMGFMLEAQP
INGANETIVSSLENLMRTKLPREVPVAVHLVSSHLVGQDIEYGLRELSWTGEKADTFNAITRAYYLRAAE
EGFPLPDHLNLPLTLRNYRVYISCCVPRKRGSKASVMEMENQVKIIRASLQGAQIATRSVDADAFIRIAG
EMINHNPDALYPTERELDPLKDLNYQCVDDTFDLKVKADYMTLGLRGNGRNSTARIMNFQLTKNPELAFL
WNMADNYSNLLNPELSVSCPFILTLVLSVEDQVKAQNEANLKFMDLEKKSKTSYAKFFPGVAQEAKEWGD
LRQRLSSGQSSLVSYFLNVTLFCRDDQETALKVEQDVLNSFRKNGFELISPRFNHMRNFLVNLPFMAGGG
LFKQLRESGVLQRAESFNVANLMPVVADSPLATSGLLAPTYRNQLAFIDLFSRGMGNTNFNMAVCGTSGA
GKTGLIQPLIRSVIDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLKFNPFANVTDDTIDEMAERLRDQL
SVMASPNGNLDEVHEGLLLKAVKATWLTKKNQARIDDVVDYLMMARDSEEYSGSPTIRSRLDEMVVLLTQ
YTRDGVYGSYFNSDEPSLKDEARMVVLELGGLEDRPSLLVAVMFSLILYIENRMYRTPRSLKKLNVIDEG
WRLLDFKNNKVGAFIEKGYRTARRHTGSYITITQNIVDFDSETASSAARAAWGNSSYKIILKQSAKEFAK
YNQLYPDQYSQLQRDMIKGFGSAKDNWFSSFMLSVEHLTSWHRLFVDPLSRAMYSSDGPDFEFLQQRRRE
GLSIHEAVWELAWKKSGPEMAALERWLHEHEGKTIS
MSNPLDSVTRTVNSLLTALKMPDASSEANDVLGQMRFPQFSRVLPYRDYDAETGLFMNEKTMGFMLEAQP
INGANETIVSSLENLMRTKLPREVPVAVHLVSSHLVGQDIEYGLRELSWTGEKADTFNAITRAYYLRAAE
EGFPLPDHLNLPLTLRNYRVYISCCVPRKRGSKASVMEMENQVKIIRASLQGAQIATRSVDADAFIRIAG
EMINHNPDALYPTERELDPLKDLNYQCVDDTFDLKVKADYMTLGLRGNGRNSTARIMNFQLTKNPELAFL
WNMADNYSNLLNPELSVSCPFILTLVLSVEDQVKAQNEANLKFMDLEKKSKTSYAKFFPGVAQEAKEWGD
LRQRLSSGQSSLVSYFLNVTLFCRDDQETALKVEQDVLNSFRKNGFELISPRFNHMRNFLVNLPFMAGGG
LFKQLRESGVLQRAESFNVANLMPVVADSPLATSGLLAPTYRNQLAFIDLFSRGMGNTNFNMAVCGTSGA
GKTGLIQPLIRSVIDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLKFNPFANVTDDTIDEMAERLRDQL
SVMASPNGNLDEVHEGLLLKAVKATWLTKKNQARIDDVVDYLMMARDSEEYSGSPTIRSRLDEMVVLLTQ
YTRDGVYGSYFNSDEPSLKDEARMVVLELGGLEDRPSLLVAVMFSLILYIENRMYRTPRSLKKLNVIDEG
WRLLDFKNNKVGAFIEKGYRTARRHTGSYITITQNIVDFDSETASSAARAAWGNSSYKIILKQSAKEFAK
YNQLYPDQYSQLQRDMIKGFGSAKDNWFSSFMLSVEHLTSWHRLFVDPLSRAMYSSDGPDFEFLQQRRRE
GLSIHEAVWELAWKKSGPEMAALERWLHEHEGKTIS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 161611..186745
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| IBG19_RS24805 | 161611..163821 | - | 2211 | WP_022652256 | type IV conjugative transfer system coupling protein TraD | virb4 |
| IBG19_RS24810 | 163955..164629 | - | 675 | WP_008324909 | tetratricopeptide repeat protein | - |
| IBG19_RS24815 | 164704..165135 | - | 432 | WP_008324118 | entry exclusion protein | - |
| IBG19_RS24820 | 165157..167979 | - | 2823 | WP_008324119 | conjugal transfer mating-pair stabilization protein TraG | traG |
| IBG19_RS24825 | 167979..169349 | - | 1371 | WP_016247527 | conjugal transfer pilus assembly protein TraH | traH |
| IBG19_RS24830 | 169336..169764 | - | 429 | WP_008324122 | hypothetical protein | - |
| IBG19_RS24835 | 169757..170311 | - | 555 | WP_008324125 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| IBG19_RS24840 | 170301..170549 | - | 249 | WP_008324126 | type-F conjugative transfer system pilin chaperone TraQ | - |
| IBG19_RS24845 | 170566..171315 | - | 750 | WP_008324127 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| IBG19_RS24850 | 171346..171534 | - | 189 | WP_008324128 | conjugal transfer protein TrbE | - |
| IBG19_RS24855 | 171566..173419 | - | 1854 | WP_022652255 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| IBG19_RS24860 | 173416..174045 | - | 630 | WP_008324130 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| IBG19_RS24865 | 174062..175048 | - | 987 | WP_032160671 | conjugal transfer pilus assembly protein TraU | traU |
| IBG19_RS24870 | 175051..175698 | - | 648 | WP_008324132 | type-F conjugative transfer system protein TraW | traW |
| IBG19_RS24875 | 175698..176045 | - | 348 | WP_008324133 | type-F conjugative transfer system protein TrbI | - |
| IBG19_RS24880 | 176042..178672 | - | 2631 | WP_008324134 | type IV secretion system protein TraC | virb4 |
| IBG19_RS24885 | 178665..179231 | - | 567 | WP_008324135 | conjugal transfer pilus-stabilizing protein TraP | - |
| IBG19_RS24890 | 179221..179622 | - | 402 | WP_008324137 | hypothetical protein | - |
| IBG19_RS24895 | 179606..179791 | - | 186 | WP_022652254 | hypothetical protein | - |
| IBG19_RS24900 | 179795..180019 | - | 225 | WP_008324141 | TraR/DksA C4-type zinc finger protein | - |
| IBG19_RS24905 | 180139..180678 | - | 540 | WP_008324154 | type IV conjugative transfer system lipoprotein TraV | traV |
| IBG19_RS24910 | 180700..182094 | - | 1395 | WP_008324156 | F-type conjugal transfer pilus assembly protein TraB | traB |
| IBG19_RS24915 | 182081..182824 | - | 744 | WP_008324157 | type-F conjugative transfer system secretin TraK | traK |
| IBG19_RS24920 | 182811..183377 | - | 567 | WP_008324158 | type IV conjugative transfer system protein TraE | traE |
| IBG19_RS24925 | 183392..183697 | - | 306 | WP_008324160 | type IV conjugative transfer system protein TraL | traL |
| IBG19_RS24930 | 183848..184198 | - | 351 | WP_008324162 | type IV conjugative transfer system pilin TraA | - |
| IBG19_RS24935 | 184256..184480 | - | 225 | WP_008324165 | TraY domain-containing protein | - |
| IBG19_RS24940 | 184567..185256 | - | 690 | WP_008324166 | hypothetical protein | - |
| IBG19_RS24945 | 185445..185837 | - | 393 | WP_008324167 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| IBG19_RS24950 | 186269..186745 | + | 477 | WP_008324168 | transglycosylase SLT domain-containing protein | virB1 |
| IBG19_RS24955 | 186792..187322 | - | 531 | WP_008324170 | antirestriction protein | - |
| IBG19_RS24960 | 187949..188779 | - | 831 | WP_022652253 | N-6 DNA methylase | - |
| IBG19_RS24965 | 188822..189085 | - | 264 | WP_008324174 | hypothetical protein | - |
| IBG19_RS24970 | 189171..189515 | - | 345 | WP_022652252 | hypothetical protein | - |
| IBG19_RS24975 | 189562..189849 | - | 288 | WP_008324178 | hypothetical protein | - |
| IBG19_RS24980 | 189903..190277 | - | 375 | WP_008324180 | hypothetical protein | - |
| IBG19_RS24985 | 190998..191264 | - | 267 | WP_022652251 | hypothetical protein | - |
| IBG19_RS24990 | 191326..191649 | - | 324 | WP_008324183 | hypothetical protein | - |
Host bacterium
| ID | 15090 | GenBank | NZ_CP072954 |
| Plasmid name | MGH 3|unnamed1 | Incompatibility group | IncFIB |
| Plasmid size | 241263 bp | Coordinate of oriT [Strand] | 186095..186197 [-] |
| Host baterium | Enterobacter sp. MGH 3 |
Cargo genes
| Drug resistance gene | tet(D), sul1, qacE, ere(A), aac(6')-IIc, sul2, blaTEM-1A, aph(3')-Ia |
| Virulence gene | - |
| Metal resistance gene | pbrA, merE, merD, merA, merC, merP, merT, merR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |