Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114634 |
| Name | oriT_pDCNB |
| Organism | Diaphorobacter sp. JS3050 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP053554 (29115..29211 [-], 97 nt) |
| oriT length | 97 nt |
| IRs (inverted repeats) | 24..31, 34..41 (TAACCGGC..GCCGGTTA) |
| Location of nic site | 60..61 |
| Conserved sequence flanking the nic site |
TCCTGCCCGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 97 nt
>oriT_pDCNB
GAATAAGGGACAGCGAAGATAGATAACCGGCCCGCCGGTTAGCTAACTTCGCACATCCTGCCCGCCTTACGGCGTTGATAACACCAAGGAAAGTCTA
GAATAAGGGACAGCGAAGATAGATAACCGGCCCGCCGGTTAGCTAACTTCGCACATCCTGCCCGCCTTACGGCGTTGATAACACCAAGGAAAGTCTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file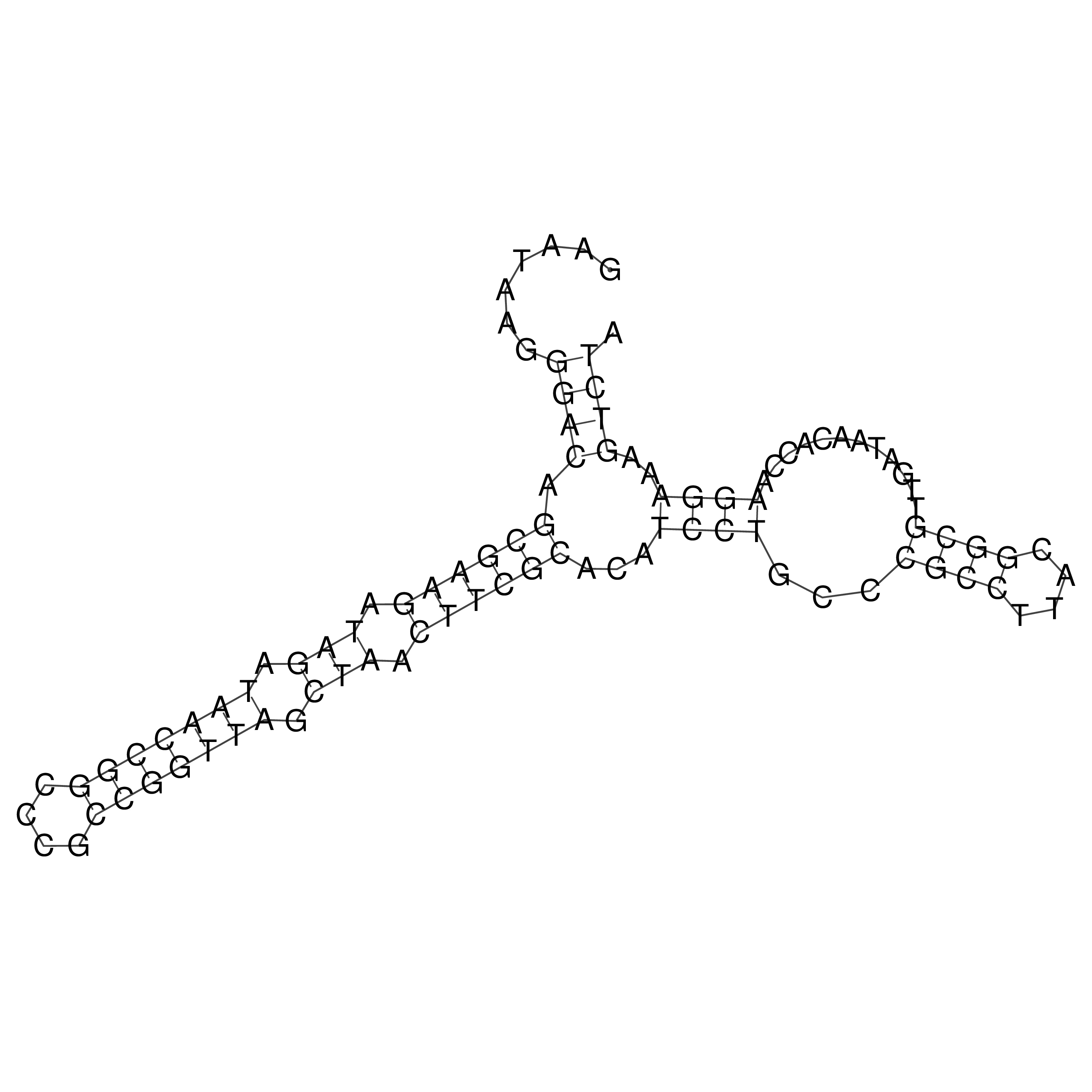
Relaxase
| ID | 9386 | GenBank | WP_012478238 |
| Name | traI_HND92_RS19080_pDCNB |
UniProt ID | A0A1H7CDK8 |
| Length | 752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 752 a.a. Molecular weight: 82867.55 Da Isoelectric Point: 10.6815
>WP_012478238.1 MULTISPECIES: TraI/MobA(P) family conjugative relaxase [Pseudomonadota]
MIAKHVPMRSLGKSDFAGLVNYVTDAQSKDHRLGQVQITNCDAVSVQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICAGLGYGEHQRVSAVHNDTDNLHIHIAINKIHPTRNTMHEPYYSHRA
LAELCTALERDYGLERDNHEPRQRGAEGRAADMEQHAGVESLVGWIKRECLDEIKGAQSWQELHQVMRDN
GLELRERANGLVIEAGDGTMVKASTLARDLSKPKLEARLGPFEASPERQTRTKAKRQYQKDPIRLRVNTV
ELYARYKAEQQTLTATRGQALEQARRRKDRLVEAAKRSGRLRRATIKVIGESGPNKKLLYAQASKALRDE
IQAINKQYQQDRQALYDSHSRRTWADWLKKEATHGNAEALTALRAREAAQGLKGNTIQGRGDPRPGHAPV
TDNITKKGTIIFRAGLTAVRDDGDKLQVSREATREGLQEALRLAMERYGSRITVNGTTQFQAQIIRAAVD
SQLPITFADPALESRRLALLKKENTHERPDRTERADQRPAEHRGRAGRGPGGPGQRAATHDDAARPIAVA
RAGFGRPGGGRAGTAGVHAGAVHRKPDIGRIGRVPPPQSQHRLRTLSQLGVVRIAGGSEVLLPRDVPRHL
EQQGTQPDHQLRRDISRPGSGVAAAPPGLAAADKYIAERESKRSKGFDIPKHARYTAGEGGLAFEGVRNI
EGQALALLKRGDEVMVMPIDQATARRMSRIAVGDAVTITPKGSIKTSKGRSR
MIAKHVPMRSLGKSDFAGLVNYVTDAQSKDHRLGQVQITNCDAVSVQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICAGLGYGEHQRVSAVHNDTDNLHIHIAINKIHPTRNTMHEPYYSHRA
LAELCTALERDYGLERDNHEPRQRGAEGRAADMEQHAGVESLVGWIKRECLDEIKGAQSWQELHQVMRDN
GLELRERANGLVIEAGDGTMVKASTLARDLSKPKLEARLGPFEASPERQTRTKAKRQYQKDPIRLRVNTV
ELYARYKAEQQTLTATRGQALEQARRRKDRLVEAAKRSGRLRRATIKVIGESGPNKKLLYAQASKALRDE
IQAINKQYQQDRQALYDSHSRRTWADWLKKEATHGNAEALTALRAREAAQGLKGNTIQGRGDPRPGHAPV
TDNITKKGTIIFRAGLTAVRDDGDKLQVSREATREGLQEALRLAMERYGSRITVNGTTQFQAQIIRAAVD
SQLPITFADPALESRRLALLKKENTHERPDRTERADQRPAEHRGRAGRGPGGPGQRAATHDDAARPIAVA
RAGFGRPGGGRAGTAGVHAGAVHRKPDIGRIGRVPPPQSQHRLRTLSQLGVVRIAGGSEVLLPRDVPRHL
EQQGTQPDHQLRRDISRPGSGVAAAPPGLAAADKYIAERESKRSKGFDIPKHARYTAGEGGLAFEGVRNI
EGQALALLKRGDEVMVMPIDQATARRMSRIAVGDAVTITPKGSIKTSKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A1H7CDK8 |
Auxiliary protein
| ID | 5507 | GenBank | WP_011255188 |
| Name | WP_011255188_pDCNB |
UniProt ID | A0A1H7CD62 |
| Length | 123 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 123 a.a. Molecular weight: 13964.97 Da Isoelectric Point: 8.4531
>WP_011255188.1 MULTISPECIES: conjugal transfer transcriptional regulator TraJ [Pseudomonadota]
MEADEQPTAKRRQHLRVPVFPDEKEQIEANAKRAGVSVARYLRDVGQGYQIRGVMDYEYVRELVRVNGDL
GRLGGLLKLWLTDDPRTARFGDATILALLGRIEATQDEMSRLMKSVVQPRAEP
MEADEQPTAKRRQHLRVPVFPDEKEQIEANAKRAGVSVARYLRDVGQGYQIRGVMDYEYVRELVRVNGDL
GRLGGLLKLWLTDDPRTARFGDATILALLGRIEATQDEMSRLMKSVVQPRAEP
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A1H7CD62 |
T4CP
| ID | 10895 | GenBank | WP_012478237 |
| Name | t4cp2_HND92_RS19085_pDCNB |
UniProt ID | _ |
| Length | 634 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 634 a.a. Molecular weight: 69616.60 Da Isoelectric Point: 8.4953
>WP_012478237.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Pseudomonadota]
MKIKMNNAVGPQVRSAKPKPSKLLPILGGVSLVGGLQAATQFFAYTFQYHASLGANLGHVYAPWSILNWS
AKWYSQYPDEIMRAGSIGMVTATVGLLGVAVAKVVTSNSSKANEYLHGSARWAEKKDIQAAGLLPRERNV
LEIVTGKDAPTATGVYVGGWQDKDGNFYYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASSSGGVCWNPLDEIRVGTEAEVGDVQNLATLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKARNDGGTATLPSVDAMLADPNRDIGELWMEMTTYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLALYRDPVVARSVSRSDFRIKDLMHSDDPVSLYIVTQPNDKARLRPLVRVMVN
MIVRLLADKMDFENGRPVAHYKHRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLRSRET
GYGHDESITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTYQEVQRPLLT
PDECLRMPGPKKNDKGEIEEAGDMVIYVAGYPAIYGKQPLYFKDPVFQARASIPAPKATDRLRQVVEAGE
GITI
MKIKMNNAVGPQVRSAKPKPSKLLPILGGVSLVGGLQAATQFFAYTFQYHASLGANLGHVYAPWSILNWS
AKWYSQYPDEIMRAGSIGMVTATVGLLGVAVAKVVTSNSSKANEYLHGSARWAEKKDIQAAGLLPRERNV
LEIVTGKDAPTATGVYVGGWQDKDGNFYYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASSSGGVCWNPLDEIRVGTEAEVGDVQNLATLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKARNDGGTATLPSVDAMLADPNRDIGELWMEMTTYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLALYRDPVVARSVSRSDFRIKDLMHSDDPVSLYIVTQPNDKARLRPLVRVMVN
MIVRLLADKMDFENGRPVAHYKHRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLRSRET
GYGHDESITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTYQEVQRPLLT
PDECLRMPGPKKNDKGEIEEAGDMVIYVAGYPAIYGKQPLYFKDPVFQARASIPAPKATDRLRQVVEAGE
GITI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 2536..14484
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HND92_RS18880 (HND92_18910) | 308..1006 | - | 699 | WP_012478194 | TraX family protein | - |
| HND92_RS18885 (HND92_18915) | 1003..1272 | - | 270 | WP_011255140 | hypothetical protein | - |
| HND92_RS18890 (HND92_18920) | 1302..1937 | - | 636 | WP_011255139 | transglycosylase SLT domain-containing protein | - |
| HND92_RS18895 (HND92_18925) | 1939..2517 | - | 579 | WP_011255138 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| HND92_RS18900 (HND92_18930) | 2536..4254 | - | 1719 | WP_012478193 | P-type conjugative transfer protein TrbL | virB6 |
| HND92_RS18905 (HND92_18935) | 4266..4487 | - | 222 | WP_011255136 | entry exclusion lipoprotein TrbK | - |
| HND92_RS18910 (HND92_18940) | 4497..5270 | - | 774 | WP_011255135 | P-type conjugative transfer protein TrbJ | virB5 |
| HND92_RS18915 (HND92_18945) | 5287..6666 | - | 1380 | WP_012478192 | TrbI/VirB10 family protein | virB10 |
| HND92_RS18920 (HND92_18950) | 6670..7131 | - | 462 | WP_011013267 | lipoprotein | - |
| HND92_RS18925 (HND92_18955) | 7134..8027 | - | 894 | WP_014386236 | P-type conjugative transfer protein TrbG | virB9 |
| HND92_RS18930 (HND92_18960) | 8045..8833 | - | 789 | WP_011013265 | conjugal transfer protein TrbF | virB8 |
| HND92_RS18935 (HND92_18965) | 8830..9348 | - | 519 | Protein_12 | conjugal transfer protein TrbE | - |
| HND92_RS18940 (HND92_18970) | 9421..10509 | + | 1089 | WP_011803559 | IS5-like element ISDisp1 family transposase | - |
| HND92_RS18945 (HND92_18975) | 10500..12713 | - | 2214 | WP_253940162 | conjugal transfer protein TrbE | virb4 |
| HND92_RS18950 (HND92_18980) | 12710..13021 | - | 312 | WP_011013263 | conjugal transfer protein TrbD | virB3 |
| HND92_RS18955 (HND92_18985) | 13025..13489 | - | 465 | WP_011013262 | conjugal transfer system pilin TrbC | virB2 |
| HND92_RS18960 (HND92_18990) | 13519..14484 | - | 966 | WP_011013261 | P-type conjugative transfer ATPase TrbB | virB11 |
| HND92_RS18965 (HND92_18995) | 14799..15158 | - | 360 | WP_011013260 | transcriptional regulator | - |
| HND92_RS18970 (HND92_19000) | 15268..15606 | + | 339 | WP_011013259 | single-stranded DNA-binding protein | - |
| HND92_RS18975 (HND92_19005) | 15655..16878 | + | 1224 | WP_011013257 | plasmid replication initiator TrfA | - |
| HND92_RS18980 (HND92_19010) | 18264..18545 | - | 282 | WP_011114071 | type II toxin-antitoxin system YafQ family toxin | - |
| HND92_RS18985 (HND92_19015) | 18532..18795 | - | 264 | WP_011114070 | type II toxin-antitoxin system RelB/DinJ family antitoxin | - |
| HND92_RS18990 (HND92_19020) | 18968..19396 | + | 429 | WP_011114069 | antirestriction protein | - |
Host bacterium
| ID | 15069 | GenBank | NZ_CP053554 |
| Plasmid name | pDCNB | Incompatibility group | - |
| Plasmid size | 49405 bp | Coordinate of oriT [Strand] | 29115..29211 [-] |
| Host baterium | Diaphorobacter sp. JS3050 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |