Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114587 |
| Name | oriT_pC45-p3 |
| Organism | Enterobacter cloacae complex sp. isolate C45 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_LT991960 (26951..26999 [-], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_pC45-p3
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file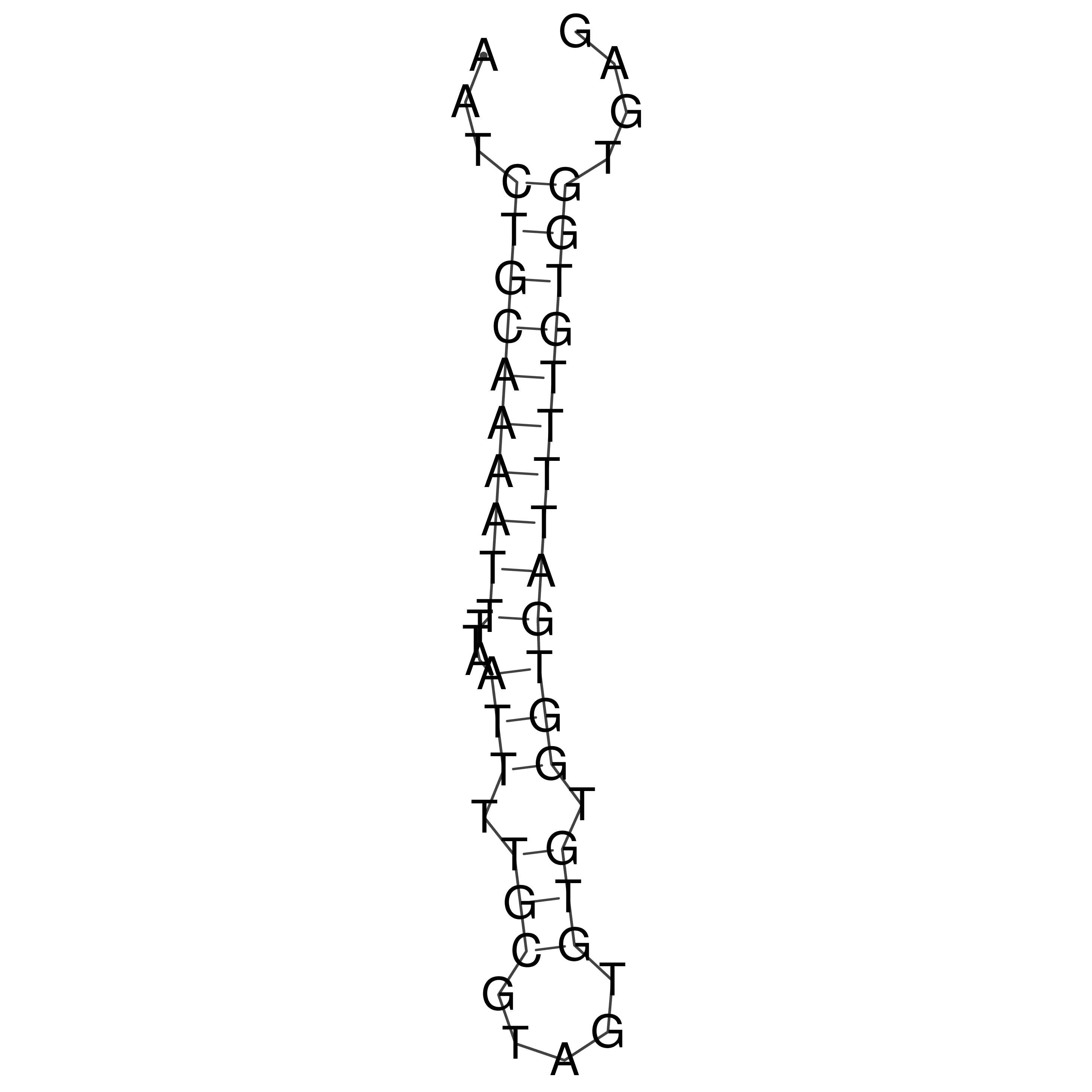
Relaxase
| ID | 9349 | GenBank | WP_107535770 |
| Name | traI_DBR02_RS26490_pC45-p3 |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190326.02 Da Isoelectric Point: 6.2242
>WP_107535770.1 conjugative transfer relaxase/helicase TraI [Enterobacter cloacae complex sp.]
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAEALGLEGKVDKQIFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVQKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSENILANRIAFGKIYQNTLRADVES
LGYRTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELTRAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGLESVAQISGTREQSVLNGLIRDSLRNEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTGSSNMLTLKDREGVRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEDGQLTVQRPGQKTTQTLSVGAGVFDGIK
IGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKARSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQIPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPTQEKAIQK
ALSEGKTLPAGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSDGKERYISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVAGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILEP
RNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAITGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKKAEQTAREMAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAEALGLEGKVDKQIFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVQKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSENILANRIAFGKIYQNTLRADVES
LGYRTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELTRAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGLESVAQISGTREQSVLNGLIRDSLRNEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTGSSNMLTLKDREGVRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEDGQLTVQRPGQKTTQTLSVGAGVFDGIK
IGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKARSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQIPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPTQEKAIQK
ALSEGKTLPAGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSDGKERYISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVAGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILEP
RNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAITGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKKAEQTAREMAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 5466 | GenBank | WP_004197733 |
| Name | WP_004197733_pC45-p3 |
UniProt ID | A0A0E3KB00 |
| Length | 129 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 129 a.a. Molecular weight: 14835.90 Da Isoelectric Point: 4.6012
>WP_004197733.1 MULTISPECIES: conjugal transfer relaxosome DNA-binding protein TraM [Bacteria]
MGKVQAYPSDEVCEKINAIVEKRRMEGAKDKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESVVKTQFSVNKILGIECLSPHVKDDPRWQWKSMVLNIQEDVQEAMRNFFPDEDAEGE
MGKVQAYPSDEVCEKINAIVEKRRMEGAKDKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESVVKTQFSVNKILGIECLSPHVKDDPRWQWKSMVLNIQEDVQEAMRNFFPDEDAEGE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A0E3KB00 |
T4CP
| ID | 10847 | GenBank | WP_064343438 |
| Name | traD_DBR02_RS26485_pC45-p3 |
UniProt ID | _ |
| Length | 769 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 769 a.a. Molecular weight: 85584.66 Da Isoelectric Point: 5.3307
>WP_064343438.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPDDAGSHAGEQPEPVSQPA
PAVMTVTPAPVKSPPTTKRPAAEPSARTAEPPVLRVTTVPLIKPKAAAAAGTAATASSAGTPAAAAGGTE
QVLAQQSAEQGQAMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDLEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPDDAGSHAGEQPEPVSQPA
PAVMTVTPAPVKSPPTTKRPAAEPSARTAEPPVLRVTTVPLIKPKAAAAAGTAATASSAGTPAAAAGGTE
QVLAQQSAEQGQAMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDLEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 29413..54521
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| DBR02_RS26285 | 24599..24946 | + | 348 | WP_022644944 | hypothetical protein | - |
| DBR02_RS26290 | 25039..25185 | + | 147 | WP_022644943 | hypothetical protein | - |
| DBR02_RS27105 | 25233..25469 | + | 237 | Protein_35 | SAM-dependent DNA methyltransferase | - |
| DBR02_RS26300 | 25818..26360 | + | 543 | WP_022644941 | antirestriction protein | - |
| DBR02_RS26305 | 26384..26806 | - | 423 | WP_072205880 | transglycosylase SLT domain-containing protein | - |
| DBR02_RS26310 | 27311..27700 | + | 390 | WP_004197733 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| DBR02_RS26315 | 27939..28613 | + | 675 | WP_004197737 | hypothetical protein | - |
| DBR02_RS26320 | 28805..28969 | + | 165 | WP_032444506 | TraY domain-containing protein | - |
| DBR02_RS26325 | 29031..29399 | + | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| DBR02_RS26330 | 29413..29718 | + | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| DBR02_RS26335 | 29738..30304 | + | 567 | WP_004194424 | type IV conjugative transfer system protein TraE | traE |
| DBR02_RS26340 | 30291..31031 | + | 741 | WP_009309865 | type-F conjugative transfer system secretin TraK | traK |
| DBR02_RS26345 | 31031..32455 | + | 1425 | WP_004194260 | F-type conjugal transfer pilus assembly protein TraB | traB |
| DBR02_RS26350 | 32569..33153 | + | 585 | WP_009309868 | type IV conjugative transfer system lipoprotein TraV | traV |
| DBR02_RS26355 | 33285..33695 | + | 411 | WP_009309869 | hypothetical protein | - |
| DBR02_RS26360 | 33700..33984 | + | 285 | WP_107535765 | hypothetical protein | - |
| DBR02_RS26365 | 34008..34232 | + | 225 | WP_060598869 | hypothetical protein | - |
| DBR02_RS26370 | 34225..34536 | + | 312 | WP_107535766 | hypothetical protein | - |
| DBR02_RS26375 | 34603..35007 | + | 405 | WP_004197817 | hypothetical protein | - |
| DBR02_RS26390 | 35384..35782 | + | 399 | WP_023179972 | hypothetical protein | - |
| DBR02_RS26395 | 35854..38493 | + | 2640 | WP_107535767 | type IV secretion system protein TraC | virb4 |
| DBR02_RS26400 | 38493..38882 | + | 390 | WP_004197815 | type-F conjugative transfer system protein TrbI | - |
| DBR02_RS26405 | 38882..39508 | + | 627 | WP_009309871 | type-F conjugative transfer system protein TraW | traW |
| DBR02_RS26410 | 39550..39939 | + | 390 | WP_004194992 | hypothetical protein | - |
| DBR02_RS26415 | 39936..40925 | + | 990 | WP_009309872 | conjugal transfer pilus assembly protein TraU | traU |
| DBR02_RS26420 | 40938..41576 | + | 639 | WP_011977786 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| DBR02_RS26425 | 41635..43590 | + | 1956 | WP_107535768 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| DBR02_RS26430 | 43622..43876 | + | 255 | WP_004195500 | conjugal transfer protein TrbE | - |
| DBR02_RS26435 | 43854..44102 | + | 249 | WP_004152675 | hypothetical protein | - |
| DBR02_RS26440 | 44115..44441 | + | 327 | WP_004194451 | hypothetical protein | - |
| DBR02_RS26445 | 44462..45214 | + | 753 | WP_042946327 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| DBR02_RS26450 | 45225..45464 | + | 240 | WP_009309875 | type-F conjugative transfer system pilin chaperone TraQ | - |
| DBR02_RS26455 | 45436..46008 | + | 573 | WP_009309876 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| DBR02_RS26460 | 46001..46429 | + | 429 | WP_004152689 | hypothetical protein | - |
| DBR02_RS26465 | 46416..47786 | + | 1371 | WP_009309877 | conjugal transfer pilus assembly protein TraH | traH |
| DBR02_RS26470 | 47786..50659 | + | 2874 | WP_107535769 | conjugal transfer mating-pair stabilization protein TraG | traG |
| DBR02_RS26750 | 51394..52086 | + | 693 | WP_009309879 | hypothetical protein | - |
| DBR02_RS26485 | 52212..54521 | + | 2310 | WP_064343438 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 15022 | GenBank | NZ_LT991960 |
| Plasmid name | pC45-p3 | Incompatibility group | - |
| Plasmid size | 65087 bp | Coordinate of oriT [Strand] | 26951..26999 [-] |
| Host baterium | Enterobacter cloacae complex sp. isolate C45 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |