Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114586 |
| Name | oriT_pC45-p2 |
| Organism | Enterobacter cloacae complex sp. isolate C45 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_LT991959 (105677..105787 [-], 111 nt) |
| oriT length | 111 nt |
| IRs (inverted repeats) | 88..93, 95..100 (AAATAA..TTATTT) 79..84, 94..99 (AATAAT..ATTATT) 27..32, 39..44 (GCAAAA..TTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 111 nt
>oriT_pC45-p2
CTCACCACAAAAGCACCACACCCCACGCAAAACGTACTTTTTGCTGATTGGCTATTCAGTTCAGTAACTTAGCGTTTGAATAATGTGAAATAAATTATTTTAAATTACTAA
CTCACCACAAAAGCACCACACCCCACGCAAAACGTACTTTTTGCTGATTGGCTATTCAGTTCAGTAACTTAGCGTTTGAATAATGTGAAATAAATTATTTTAAATTACTAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file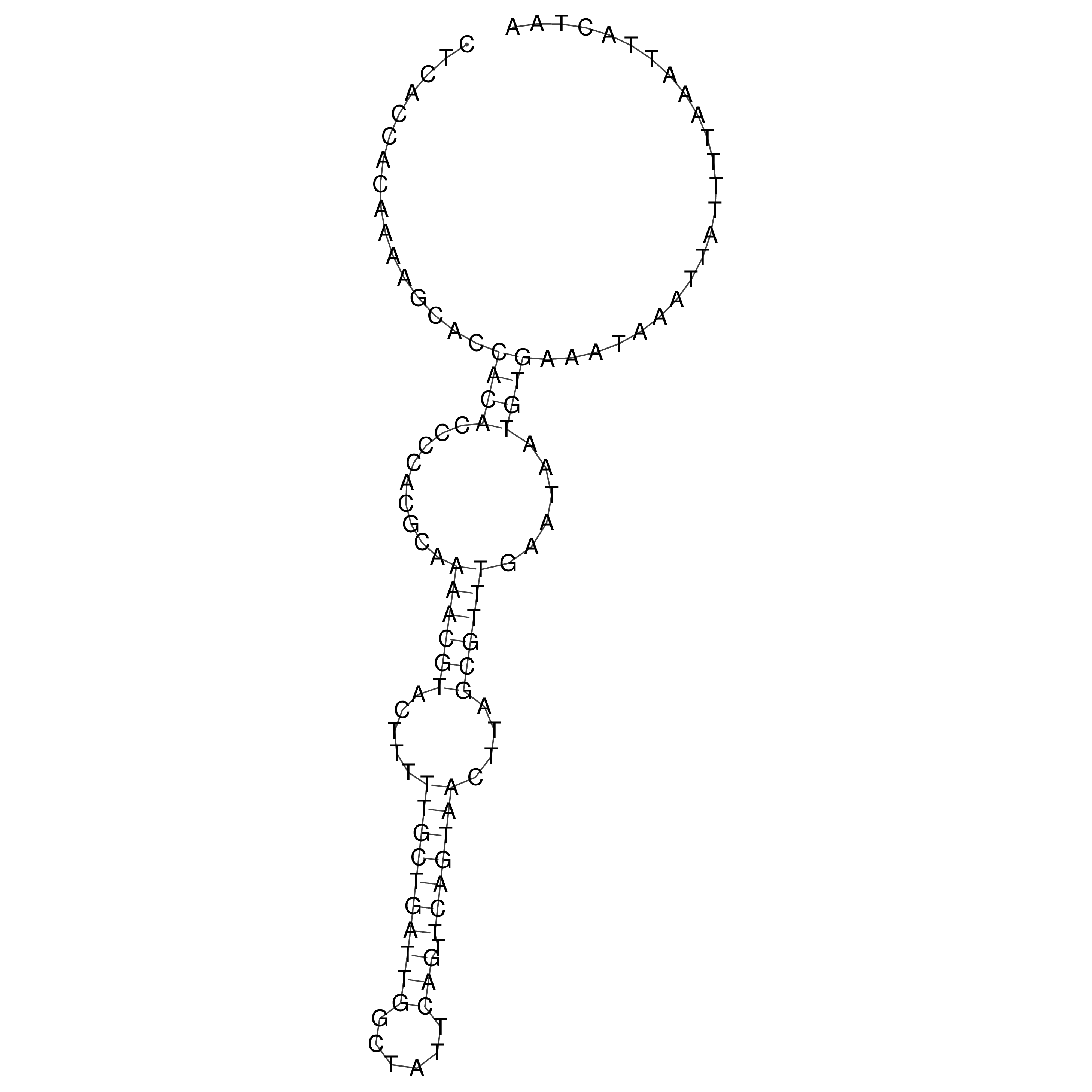
Relaxase
| ID | 9348 | GenBank | WP_107535705 |
| Name | traI_DBR02_RS25710_pC45-p2 |
UniProt ID | _ |
| Length | 1747 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1747 a.a. Molecular weight: 189239.45 Da Isoelectric Point: 6.6854
>WP_107535705.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacter]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGEGAKALGLSGNVDVKTFTRVLEGKLPDGSDLSRAQ
DGTNKHRPGYDLTFSAPKSISVMAMLGGDTRLIDAHNRAVDTAIKQIEALASTRVMTDGKSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHDGTWRTLSSDKIGKTGFIENVYANQIAFGRLYRAALKDDVTS
MGYETETVGKHGMWELKGVPTEPYSSRSKAIREAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMAT
LKETGFDIQAYREAADLRVREGAVPVSTPESPDAPAAVSQSIAMLSDRRARFTYSELLATTLGQLPARPG
MIEQAREGIDAAIKNEQLIPLDKEKGLFTSNVHVLDELSVSALSRELQQQGRVDVFPDKSVPRSRAFSDA
VSVLAQDRSPVAIISGQGGAAGQRERVAELAMMAREQGRDVQIMAADRRSAANLARDERLSGELITDRRG
LTEGMTFIPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTRVSVISEPDRRERYDRLAADFAKSIRAGKEAVVQISGAREQAALAATVRDALKGEKVLGERDISIT
TLEPVWLDGRHRQVRDHYREGMVMERWNAEERSRERFVIDRVTARNNSLTLRNAQGETQVTRLTELDSSW
SLYRTGTLQVTEGDRLAVLGQTQGARLKGGDSVTVTGIDEKGIHVSLPGRKTQVVLPAGDSPFTAPKIGQ
GWVESPGRSVSDSATVYASLTQREMDNATLNGLARSGAEIRLYSAQTVQKTEEKLSRQSAWTVVTAQVKE
AAGKDSLGEALAHQKAALHTPEQQAIHLAIPSLEGNGLAFTRPQLMAAAKDFAQDKLSLAAIEDEIARQT
RSGALISVPVSQGNGLQQLVSRQSYNAEKSIIRHVLEGKDAVTPLMGKVPDAQLAGLTDGQQNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEERRPRVIGVAPTHRAVGEMRDAGVPKSQTLAAFIHDTQ
QQLRGGERPDFSNVLFLIDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DTAVMQEIVRQTPALRPAVYSLIERNIGSALTTLESVAPQQVPRRPDAWQPDGSAMEFSREQEKAIADAI
KAGDLAPGGQPATLLEAIVKDYTGRTPQAQAQTIVITALNADRRQLNAMIHDARREAGELGEKEAIVPVL
TPANIRDGELRRMDTWQSHASSMVLLDNTYYRIDALDKDAHLVTLKDAQGNTRSLSPAQAVTEGVTLYRQ
DTIAVSPGDRMRFSKSDNDRGFVANSVWTVSDIKGDSVTLTDGKQTRTVRPGVEPAEQHIDLAYAVTVYG
AQGASEPFSISLQGTDGGRKQMVNFESAYVALSRMKQHAQVYTDNREKWVAAMEKSQAKSTAHDIVEPRN
DRAVANAARLTASAKGLGEVPAGRAALRQAGLRPETTLAKFISPGRKYPQPHVALPAFDRNGRQAGVWLS
TLTDGDGRLQGLSGEGRVMGSGDAAFAGLQASRNGESLMAGDMAEGVRLARENPQSGVVVRIGDAEGRPW
NPGAITGGRVWADVVPDGTGTQHGEKIPPEVLAQQELEAQQRRELEKRAEDAVRELARGGEKSAEPADAV
RELARELGEGKNRERSTDVTLPESPDVRARDEAVSRVAHENVQRDRLQQMERDTVRVLEREKTLGGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGEGAKALGLSGNVDVKTFTRVLEGKLPDGSDLSRAQ
DGTNKHRPGYDLTFSAPKSISVMAMLGGDTRLIDAHNRAVDTAIKQIEALASTRVMTDGKSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHDGTWRTLSSDKIGKTGFIENVYANQIAFGRLYRAALKDDVTS
MGYETETVGKHGMWELKGVPTEPYSSRSKAIREAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMAT
LKETGFDIQAYREAADLRVREGAVPVSTPESPDAPAAVSQSIAMLSDRRARFTYSELLATTLGQLPARPG
MIEQAREGIDAAIKNEQLIPLDKEKGLFTSNVHVLDELSVSALSRELQQQGRVDVFPDKSVPRSRAFSDA
VSVLAQDRSPVAIISGQGGAAGQRERVAELAMMAREQGRDVQIMAADRRSAANLARDERLSGELITDRRG
LTEGMTFIPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTRVSVISEPDRRERYDRLAADFAKSIRAGKEAVVQISGAREQAALAATVRDALKGEKVLGERDISIT
TLEPVWLDGRHRQVRDHYREGMVMERWNAEERSRERFVIDRVTARNNSLTLRNAQGETQVTRLTELDSSW
SLYRTGTLQVTEGDRLAVLGQTQGARLKGGDSVTVTGIDEKGIHVSLPGRKTQVVLPAGDSPFTAPKIGQ
GWVESPGRSVSDSATVYASLTQREMDNATLNGLARSGAEIRLYSAQTVQKTEEKLSRQSAWTVVTAQVKE
AAGKDSLGEALAHQKAALHTPEQQAIHLAIPSLEGNGLAFTRPQLMAAAKDFAQDKLSLAAIEDEIARQT
RSGALISVPVSQGNGLQQLVSRQSYNAEKSIIRHVLEGKDAVTPLMGKVPDAQLAGLTDGQQNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEERRPRVIGVAPTHRAVGEMRDAGVPKSQTLAAFIHDTQ
QQLRGGERPDFSNVLFLIDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DTAVMQEIVRQTPALRPAVYSLIERNIGSALTTLESVAPQQVPRRPDAWQPDGSAMEFSREQEKAIADAI
KAGDLAPGGQPATLLEAIVKDYTGRTPQAQAQTIVITALNADRRQLNAMIHDARREAGELGEKEAIVPVL
TPANIRDGELRRMDTWQSHASSMVLLDNTYYRIDALDKDAHLVTLKDAQGNTRSLSPAQAVTEGVTLYRQ
DTIAVSPGDRMRFSKSDNDRGFVANSVWTVSDIKGDSVTLTDGKQTRTVRPGVEPAEQHIDLAYAVTVYG
AQGASEPFSISLQGTDGGRKQMVNFESAYVALSRMKQHAQVYTDNREKWVAAMEKSQAKSTAHDIVEPRN
DRAVANAARLTASAKGLGEVPAGRAALRQAGLRPETTLAKFISPGRKYPQPHVALPAFDRNGRQAGVWLS
TLTDGDGRLQGLSGEGRVMGSGDAAFAGLQASRNGESLMAGDMAEGVRLARENPQSGVVVRIGDAEGRPW
NPGAITGGRVWADVVPDGTGTQHGEKIPPEVLAQQELEAQQRRELEKRAEDAVRELARGGEKSAEPADAV
RELARELGEGKNRERSTDVTLPESPDVRARDEAVSRVAHENVQRDRLQQMERDTVRVLEREKTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 10846 | GenBank | WP_107535706 |
| Name | traD_DBR02_RS25715_pC45-p2 |
UniProt ID | _ |
| Length | 740 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 740 a.a. Molecular weight: 83132.85 Da Isoelectric Point: 5.0430
>WP_107535706.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacter]
MSFNAKDMTQGGQIASMRIRMFSQIANIILYCLVILFGIIVALTLTVKLSWQTFVNGMVWYWCQTLAPMR
DVLKSQPVYTIRYYGKELKYNAQQLLQDKYVIWCADQVWTAFILAAIIAAVVCVIAFFVTTWILGHQGKQ
QSEDEVTGGRQLTDDPREVARMLKRDNLASDIKIGDLPLIKNSEIQNFCLHGTVGAGKSEVIRRLMNYAR
KRGDMVIVYDRSCEFVKSYYDPAIDKILNPMDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLQNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGEPFTIRDWMRGVREDGRNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDIQTLPDLSCYVTLPGPYPAVKMAL
KYQERPKVAPEFIQRRIDDKAEQRLNAALAAREAESRSVAMLFEPDTEAIAPVSAPENAGQPQQPQQPQQ
PQQPQQPQETQPAPVITPRPAPVAASTSAAAAAAVAGTGGTESVLETKAEEAEQLPPGIDESGEVVDMAA
WEAWQAEQDELTPQERQRREEVNINVHRTHDDDLEPGGYI
MSFNAKDMTQGGQIASMRIRMFSQIANIILYCLVILFGIIVALTLTVKLSWQTFVNGMVWYWCQTLAPMR
DVLKSQPVYTIRYYGKELKYNAQQLLQDKYVIWCADQVWTAFILAAIIAAVVCVIAFFVTTWILGHQGKQ
QSEDEVTGGRQLTDDPREVARMLKRDNLASDIKIGDLPLIKNSEIQNFCLHGTVGAGKSEVIRRLMNYAR
KRGDMVIVYDRSCEFVKSYYDPAIDKILNPMDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLQNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGEPFTIRDWMRGVREDGRNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDIQTLPDLSCYVTLPGPYPAVKMAL
KYQERPKVAPEFIQRRIDDKAEQRLNAALAAREAESRSVAMLFEPDTEAIAPVSAPENAGQPQQPQQPQQ
PQQPQQPQETQPAPVITPRPAPVAASTSAAAAAAVAGTGGTESVLETKAEEAEQLPPGIDESGEVVDMAA
WEAWQAEQDELTPQERQRREEVNINVHRTHDDDLEPGGYI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 82278..106331
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| DBR02_RS25715 | 82278..84500 | - | 2223 | WP_107535706 | type IV conjugative transfer system coupling protein TraD | virb4 |
| DBR02_RS25720 | 84621..85421 | - | 801 | WP_107535707 | hypothetical protein | - |
| DBR02_RS25730 | 85967..88789 | - | 2823 | WP_107535709 | conjugal transfer mating-pair stabilization protein TraG | traG |
| DBR02_RS25735 | 88789..90159 | - | 1371 | WP_107535710 | conjugal transfer pilus assembly protein TraH | traH |
| DBR02_RS25740 | 90146..90541 | - | 396 | WP_233157809 | conjugal transfer protein TrbF | - |
| DBR02_RS25745 | 90570..91118 | - | 549 | WP_107535712 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| DBR02_RS25750 | 91108..91353 | - | 246 | WP_102750541 | type-F conjugative transfer system pilin chaperone TraQ | - |
| DBR02_RS25755 | 91370..92125 | - | 756 | WP_107535713 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| DBR02_RS25760 | 92135..93994 | - | 1860 | WP_107535714 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| DBR02_RS25765 | 93991..94572 | - | 582 | WP_233157812 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| DBR02_RS25770 | 94624..95610 | - | 987 | WP_107535716 | conjugal transfer pilus assembly protein TraU | traU |
| DBR02_RS25775 | 95607..96254 | - | 648 | WP_107535717 | type-F conjugative transfer system protein TraW | traW |
| DBR02_RS25780 | 96254..96601 | - | 348 | WP_102750535 | type-F conjugative transfer system protein TrbI | - |
| DBR02_RS25785 | 96598..99228 | - | 2631 | WP_107535718 | type IV secretion system protein TraC | virb4 |
| DBR02_RS26850 | 99225..99389 | - | 165 | WP_173937213 | hypothetical protein | - |
| DBR02_RS25790 | 99392..99622 | - | 231 | WP_107535719 | TraR/DksA C4-type zinc finger protein | - |
| DBR02_RS25795 | 99744..100280 | - | 537 | WP_107535720 | type IV conjugative transfer system lipoprotein TraV | traV |
| DBR02_RS25800 | 100302..101705 | - | 1404 | WP_107535721 | F-type conjugal transfer pilus assembly protein TraB | traB |
| DBR02_RS25805 | 101692..102435 | - | 744 | WP_107535722 | type-F conjugative transfer system secretin TraK | traK |
| DBR02_RS25810 | 102422..102988 | - | 567 | WP_063931979 | type IV conjugative transfer system protein TraE | traE |
| DBR02_RS25815 | 103003..103308 | - | 306 | WP_063449767 | type IV conjugative transfer system protein TraL | traL |
| DBR02_RS25820 | 103445..103795 | - | 351 | WP_046622497 | type IV conjugative transfer system pilin TraA | - |
| DBR02_RS25825 | 103842..104066 | - | 225 | WP_046622499 | TraY domain-containing protein | - |
| DBR02_RS25830 | 104145..104849 | - | 705 | WP_107535723 | PAS fold family protein | - |
| DBR02_RS25835 | 105033..105428 | - | 396 | WP_107535724 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| DBR02_RS25840 | 105858..106331 | + | 474 | WP_063449770 | transglycosylase SLT domain-containing protein | virB1 |
| DBR02_RS25845 | 106372..106902 | - | 531 | WP_107535725 | antirestriction protein | - |
| DBR02_RS25855 | 107563..108393 | - | 831 | WP_107535727 | N-6 DNA methylase | - |
| DBR02_RS25860 | 108439..108705 | - | 267 | WP_107535728 | hypothetical protein | - |
| DBR02_RS25865 | 108790..109146 | - | 357 | WP_107535729 | hypothetical protein | - |
| DBR02_RS25870 | 109196..109477 | - | 282 | WP_233157808 | hypothetical protein | - |
| DBR02_RS25875 | 109539..109928 | - | 390 | WP_102750519 | ammonia monooxygenase | - |
| DBR02_RS26675 | 109952..110179 | + | 228 | WP_157959564 | hypothetical protein | - |
Host bacterium
| ID | 15021 | GenBank | NZ_LT991959 |
| Plasmid name | pC45-p2 | Incompatibility group | IncFIB |
| Plasmid size | 142732 bp | Coordinate of oriT [Strand] | 105677..105787 [-] |
| Host baterium | Enterobacter cloacae complex sp. isolate C45 |
Cargo genes
| Drug resistance gene | sul1, aph(6)-Id, aph(3'')-Ib |
| Virulence gene | - |
| Metal resistance gene | silE, silS, silR, silC, silF, silB, silA, silP, pcoA, pcoB, pcoC, pcoD, pcoR, pcoS, pcoE, merR, merT, merP, merC, merA, merD, merE, arsH, arsC, arsB |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |