Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114581 |
| Name | oriT_MA01|punnamed1 |
| Organism | Agrobacterium sp. MA01 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP045855 (88231..88274 [+], 44 nt) |
| oriT length | 44 nt |
| IRs (inverted repeats) | 23..28, 33..38 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 44 nt
>oriT_MA01|punnamed1
GGATCCGAAGGGCGCAATTATACGTCGCTGATGCGACGCTCTGC
GGATCCGAAGGGCGCAATTATACGTCGCTGATGCGACGCTCTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file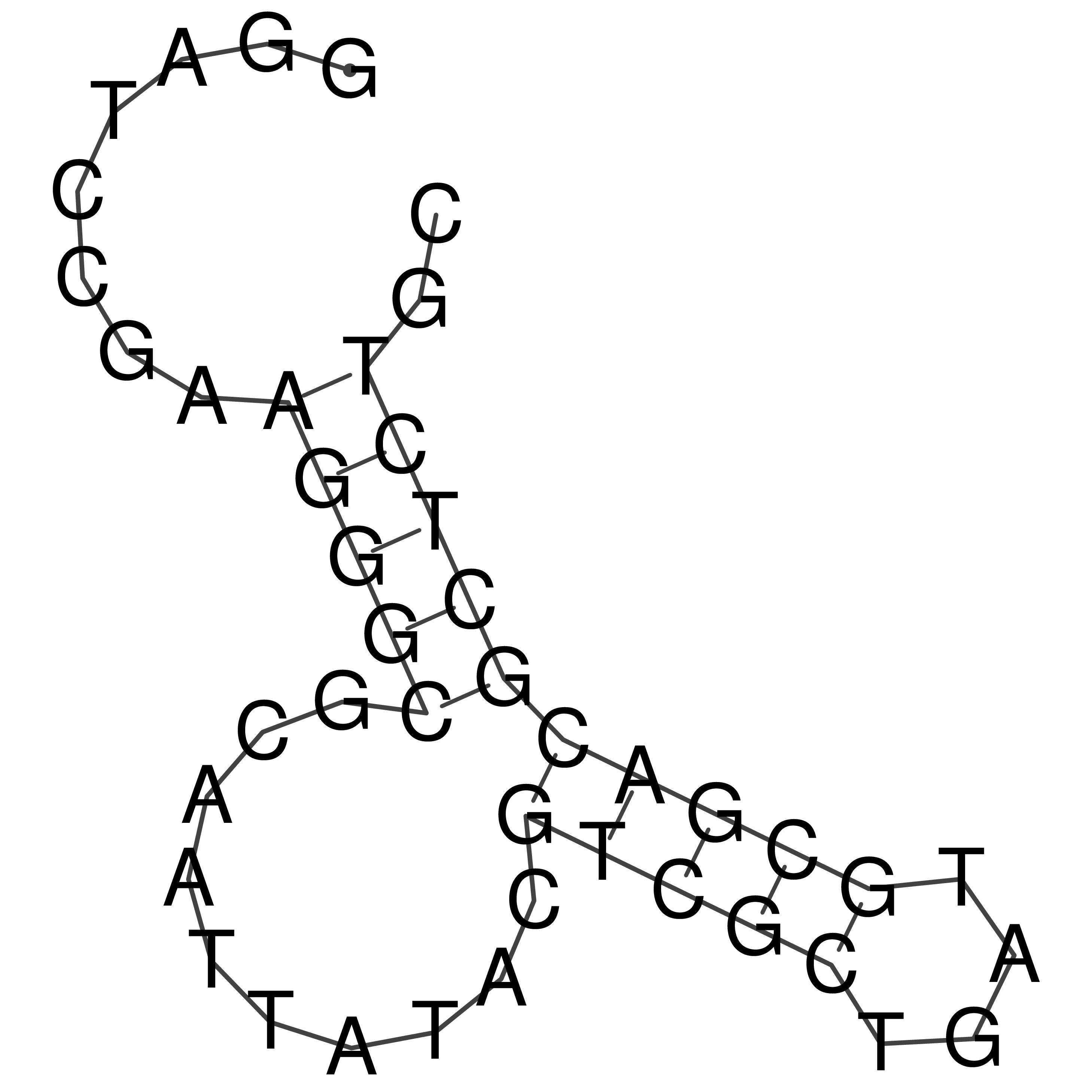
Relaxase
| ID | 9343 | GenBank | WP_153763737 |
| Name | traA_GH983_RS22435_MA01|punnamed1 |
UniProt ID | _ |
| Length | 1106 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1106 a.a. Molecular weight: 122426.18 Da Isoelectric Point: 9.4958
>WP_153763737.1 Ti-type conjugative transfer relaxase TraA [Agrobacterium sp. MA01]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMDYEREARTIDYTRKHGLLHEEFVISPDAPQWLQSMIADR
SVSGASEAFWNKVEEFERRSDAQLAKDVTIALPIELSVEQNIELVRDFVARHITAEGMVADWVFHDAPGN
PHIHLMTTLRPLTEDGFGSKKVAVTGPDGKALRNNSGKIVYELWAGGLDDFNAFRDGWFACQNRHLALAG
LDIRIDGRSFEKQGIELTPTIHLGVGTKAIERKAASGNGPAEEKAALERLELQEERRAENAQRIQRNPAI
VLDLITREKSVFDERDVAKVLHRYIDDVGLFQSLMARILQLPETLRLDRERVDFATGVRAPTKYTTRELI
RLESEMANRATWLSQRSSHGVGKTVLERTFAHHERLSDEQKVAIEHVVGVERIAAVIGRAGAGKTTMMKA
AREAWEAAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWDQGRGQLDHKTVFVLDEAGMVSSRQMA
TFVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQREQWMRDASLDLARGHVGKAL
AAYQSNGKMIGLELKADAVTNLIADWNRDYDPAKSTLILAHLRRDVRMLNELARMKLIERGIIEQGLVFK
TADGNRNFAAGDQIVFLKNEGSIGVKNGMLGKVLEATPGRLVAVIGEGEHQRHVTVEQSFYNNIDHGYAT
TVHKSQGATVDRVNVLASLSLDRHLTYVAMTRHREDLGVYYGRRSFAKAGGLVEILSRRNSKETTLDYAG
GSTYRQALRFAEAHGLHIINVARTVARDRLEWTVQQKQKLLDLAGRLAAIGAKLGFATSIANTVPTAAKE
TKPMVAGITTYPKSIDQAVEDKLAADPALKRQWEEVSMRFHHVYAQPESAFKAVDVDAMLRDQAAAAKTI
TKIASEPETFGALKGKTGLLASRADKVDRDRALKNVTPLADSISDYLRQRGDAERRNHAEELAVRRRVAL
DIPALSSNAKSVLERVRDAIDRNDLPSGLEYTLADKMVKAELEGFAKAVTERFGERTFLPLAAKDPNGEA
FLRVTSGMNTAQRSEVQQAWNTMRTVQQLSTHERSVTALKQAEALRQTRSQGLSLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMDYEREARTIDYTRKHGLLHEEFVISPDAPQWLQSMIADR
SVSGASEAFWNKVEEFERRSDAQLAKDVTIALPIELSVEQNIELVRDFVARHITAEGMVADWVFHDAPGN
PHIHLMTTLRPLTEDGFGSKKVAVTGPDGKALRNNSGKIVYELWAGGLDDFNAFRDGWFACQNRHLALAG
LDIRIDGRSFEKQGIELTPTIHLGVGTKAIERKAASGNGPAEEKAALERLELQEERRAENAQRIQRNPAI
VLDLITREKSVFDERDVAKVLHRYIDDVGLFQSLMARILQLPETLRLDRERVDFATGVRAPTKYTTRELI
RLESEMANRATWLSQRSSHGVGKTVLERTFAHHERLSDEQKVAIEHVVGVERIAAVIGRAGAGKTTMMKA
AREAWEAAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWDQGRGQLDHKTVFVLDEAGMVSSRQMA
TFVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQREQWMRDASLDLARGHVGKAL
AAYQSNGKMIGLELKADAVTNLIADWNRDYDPAKSTLILAHLRRDVRMLNELARMKLIERGIIEQGLVFK
TADGNRNFAAGDQIVFLKNEGSIGVKNGMLGKVLEATPGRLVAVIGEGEHQRHVTVEQSFYNNIDHGYAT
TVHKSQGATVDRVNVLASLSLDRHLTYVAMTRHREDLGVYYGRRSFAKAGGLVEILSRRNSKETTLDYAG
GSTYRQALRFAEAHGLHIINVARTVARDRLEWTVQQKQKLLDLAGRLAAIGAKLGFATSIANTVPTAAKE
TKPMVAGITTYPKSIDQAVEDKLAADPALKRQWEEVSMRFHHVYAQPESAFKAVDVDAMLRDQAAAAKTI
TKIASEPETFGALKGKTGLLASRADKVDRDRALKNVTPLADSISDYLRQRGDAERRNHAEELAVRRRVAL
DIPALSSNAKSVLERVRDAIDRNDLPSGLEYTLADKMVKAELEGFAKAVTERFGERTFLPLAAKDPNGEA
FLRVTSGMNTAQRSEVQQAWNTMRTVQQLSTHERSVTALKQAEALRQTRSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 10838 | GenBank | WP_153763736 |
| Name | traG_GH983_RS22420_MA01|punnamed1 |
UniProt ID | _ |
| Length | 634 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 634 a.a. Molecular weight: 68299.99 Da Isoelectric Point: 9.0918
>WP_153763736.1 Ti-type conjugative transfer system protein TraG [Agrobacterium sp. MA01]
MKVNRVVLFSLPVAMMVATVFTMTGMETWLSGFGQTEAAKIVLGRAGIAAPYVAAAALALVLLFATAGSA
SICAAGVGAAIGNAAVILIACVREGGRLAELSSQVPAGQSALSYLDPSTMIGAAAAFMSGCFAIRVAMVG
NAAFARATPTRITGKRALHGEADWMKLTEAEKLFPGSGGIVVGERYRVDRDSVAGVSFRADNGETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGSLVVLDPSNEVAPMVSKHREEAGRRVRILD
PRKPATGFNALDWVGQYGGTKEEDIASVASWIMSDSGRASGVRDDFFRASGLQLLTAIIADVCLSGHTPT
ERQTLRQVRENLSEPEPKLRQRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAIKETHWLSYQNYA
AVVSGSTFRTDDIASGKTDVFINIDLKTLETHAGLARVVIGSFLNAIYNRDGAMEGRALFLLDEIARLGY
MRILETARDAGRKYGITLTMIYQSIGQLRETYGGRDASSKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQSRGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNPPLRCGRAIWFRRKDMTTCVK
LSRF
MKVNRVVLFSLPVAMMVATVFTMTGMETWLSGFGQTEAAKIVLGRAGIAAPYVAAAALALVLLFATAGSA
SICAAGVGAAIGNAAVILIACVREGGRLAELSSQVPAGQSALSYLDPSTMIGAAAAFMSGCFAIRVAMVG
NAAFARATPTRITGKRALHGEADWMKLTEAEKLFPGSGGIVVGERYRVDRDSVAGVSFRADNGETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGSLVVLDPSNEVAPMVSKHREEAGRRVRILD
PRKPATGFNALDWVGQYGGTKEEDIASVASWIMSDSGRASGVRDDFFRASGLQLLTAIIADVCLSGHTPT
ERQTLRQVRENLSEPEPKLRQRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAIKETHWLSYQNYA
AVVSGSTFRTDDIASGKTDVFINIDLKTLETHAGLARVVIGSFLNAIYNRDGAMEGRALFLLDEIARLGY
MRILETARDAGRKYGITLTMIYQSIGQLRETYGGRDASSKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQSRGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNPPLRCGRAIWFRRKDMTTCVK
LSRF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 10839 | GenBank | WP_153763743 |
| Name | t4cp2_GH983_RS22515_MA01|punnamed1 |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 91830.54 Da Isoelectric Point: 5.8553
>WP_153763743.1 conjugal transfer protein TrbE [Agrobacterium sp. MA01]
MAALKHFRATGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAILSRLG
TGWMIQVEAIRVPTVDYPTPDQCHFPDPVTRAIDLERRAHFRREQGHFESKHAIILTYRPLESKKTALAK
YIYSDEESRKKSYADTVLSVFRNAIREIEQYLANTLSIARMQTREVRERGGERVARYDDLLQFVRFCITG
ESHPVRLPEVPMYLDWLATAELEHGLTPKVENRFLAVVAIDGLPAESWPGILNNLDLMPLTYRWSSRFIF
LDAEDARQKLERTRKKWQQKVRPFFDQLFQTQSRSADQDAMTMVAETEDAIAQASSQLVAYGYYTPVVIL
FDEERDALQEKAEAIRRLVQAEGFGARVETLNATDAFLGSLPGNWYANIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYDCAQIFA
FDKGNSLLPLTLAAGGDHYEIGNDQKGEERALAFCPLFDLSTDGDRAWATEWIETLVALQGVTITPDHRN
AISRQIGLMATAPGKSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLTLRAFQCFEIEQLMNLG
ERNLVPVLTYLFHRIEKRLDGSPSLILLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVSGAIPKREYYVATPDGRRLFDMSLGP
VALSFVGASGKEDLKRIRALKAEHGQEWPTHWLESRGVHDATSHLNVR
MAALKHFRATGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAILSRLG
TGWMIQVEAIRVPTVDYPTPDQCHFPDPVTRAIDLERRAHFRREQGHFESKHAIILTYRPLESKKTALAK
YIYSDEESRKKSYADTVLSVFRNAIREIEQYLANTLSIARMQTREVRERGGERVARYDDLLQFVRFCITG
ESHPVRLPEVPMYLDWLATAELEHGLTPKVENRFLAVVAIDGLPAESWPGILNNLDLMPLTYRWSSRFIF
LDAEDARQKLERTRKKWQQKVRPFFDQLFQTQSRSADQDAMTMVAETEDAIAQASSQLVAYGYYTPVVIL
FDEERDALQEKAEAIRRLVQAEGFGARVETLNATDAFLGSLPGNWYANIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYDCAQIFA
FDKGNSLLPLTLAAGGDHYEIGNDQKGEERALAFCPLFDLSTDGDRAWATEWIETLVALQGVTITPDHRN
AISRQIGLMATAPGKSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLTLRAFQCFEIEQLMNLG
ERNLVPVLTYLFHRIEKRLDGSPSLILLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVSGAIPKREYYVATPDGRRLFDMSLGP
VALSFVGASGKEDLKRIRALKAEHGQEWPTHWLESRGVHDATSHLNVR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 93410..106278
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| GH983_RS22440 (GH983_22440) | 91666..92232 | + | 567 | WP_097143196 | conjugative transfer signal peptidase TraF | - |
| GH983_RS22445 (GH983_22445) | 92222..93394 | + | 1173 | WP_097143197 | conjugal transfer protein TraB | - |
| GH983_RS22450 (GH983_22450) | 93410..94015 | + | 606 | WP_153763738 | TraH family protein | virB1 |
| GH983_RS22455 (GH983_22455) | 94058..94435 | - | 378 | WP_153763739 | DUF5615 family PIN-like protein | - |
| GH983_RS22460 (GH983_22460) | 94440..95063 | - | 624 | WP_153763740 | DUF433 domain-containing protein | - |
| GH983_RS22465 (GH983_22465) | 95185..95403 | - | 219 | WP_097143201 | helix-turn-helix domain-containing protein | - |
| GH983_RS22470 (GH983_22470) | 95543..95863 | + | 321 | WP_097143202 | transcriptional repressor TraM | - |
| GH983_RS22475 (GH983_22475) | 95865..96566 | - | 702 | WP_194842846 | autoinducer binding domain-containing protein | - |
| GH983_RS22480 (GH983_22480) | 96744..98081 | - | 1338 | WP_153763742 | IncP-type conjugal transfer protein TrbI | virB10 |
| GH983_RS22485 (GH983_22485) | 98096..98533 | - | 438 | WP_097143205 | conjugal transfer protein TrbH | - |
| GH983_RS22490 (GH983_22490) | 98537..99349 | - | 813 | WP_097143206 | P-type conjugative transfer protein TrbG | virB9 |
| GH983_RS22495 (GH983_22495) | 99366..100028 | - | 663 | WP_097143207 | conjugal transfer protein TrbF | virB8 |
| GH983_RS22500 (GH983_22500) | 100049..101227 | - | 1179 | WP_097143208 | P-type conjugative transfer protein TrbL | virB6 |
| GH983_RS22505 (GH983_22505) | 101221..101421 | - | 201 | WP_097143209 | entry exclusion protein TrbK | - |
| GH983_RS22510 (GH983_22510) | 101418..102176 | - | 759 | WP_246727636 | P-type conjugative transfer protein TrbJ | virB5 |
| GH983_RS22515 (GH983_22515) | 102193..104649 | - | 2457 | WP_153763743 | conjugal transfer protein TrbE | virb4 |
| GH983_RS22520 (GH983_22520) | 104659..104943 | - | 285 | WP_113426515 | conjugal transfer protein TrbD | virB3 |
| GH983_RS22525 (GH983_22525) | 104940..105317 | - | 378 | WP_097143212 | TrbC/VirB2 family protein | virB2 |
| GH983_RS22530 (GH983_22530) | 105307..106278 | - | 972 | WP_097143213 | P-type conjugative transfer ATPase TrbB | virB11 |
| GH983_RS22535 (GH983_22535) | 106281..106907 | - | 627 | WP_097143214 | acyl-homoserine-lactone synthase TraI | - |
| GH983_RS22540 (GH983_22540) | 107289..108488 | + | 1200 | WP_084438685 | plasmid partitioning protein RepA | - |
| GH983_RS22545 (GH983_22545) | 108581..109603 | + | 1023 | WP_113426518 | plasmid partitioning protein RepB | - |
| GH983_RS22550 (GH983_22550) | 109756..110994 | + | 1239 | WP_097143216 | plasmid replication protein RepC | - |
Host bacterium
| ID | 15016 | GenBank | NZ_CP045855 |
| Plasmid name | MA01|punnamed1 | Incompatibility group | - |
| Plasmid size | 132451 bp | Coordinate of oriT [Strand] | 88231..88274 [+] |
| Host baterium | Agrobacterium sp. MA01 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |