Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114428 |
| Name | oriT_pH12-1 |
| Organism | Citrobacter sp. H12-3-2 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP045838 (57652..57736 [-], 85 nt) |
| oriT length | 85 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 85 nt
>oriT_pH12-1
GGGTGTCGGGGCGGAGCCCTGACCAGGTGGTGTTTGTAATATCGTGCGCGCGCGGTATTACAAACACACATCCTGTCCCGTCTTT
GGGTGTCGGGGCGGAGCCCTGACCAGGTGGTGTTTGTAATATCGTGCGCGCGCGGTATTACAAACACACATCCTGTCCCGTCTTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file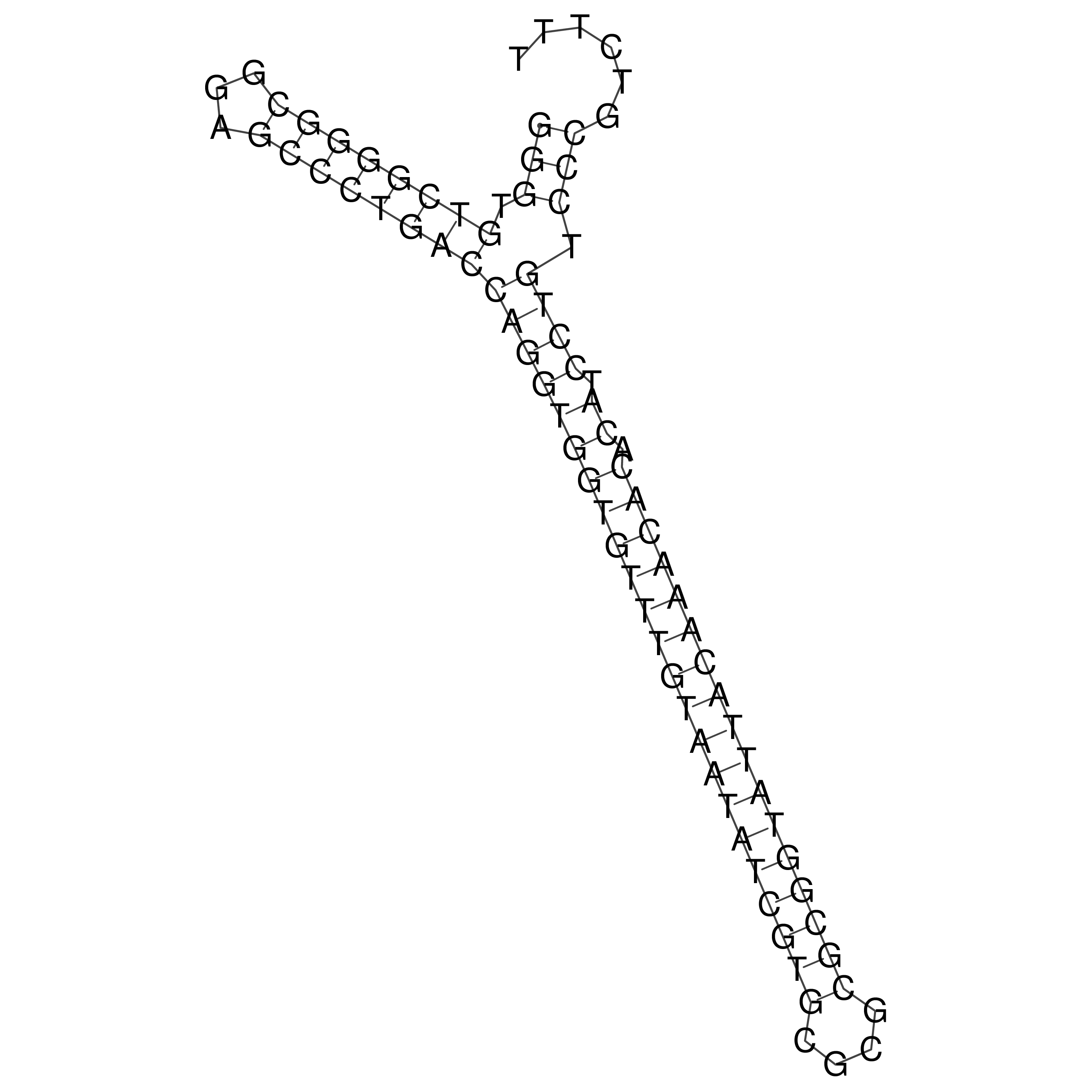
Relaxase
| ID | 9249 | GenBank | WP_153754027 |
| Name | Relaxase_GHC22_RS23820_pH12-1 |
UniProt ID | _ |
| Length | 899 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 102404.56 Da Isoelectric Point: 8.8125
>WP_153754027.1 MULTISPECIES: relaxase/mobilization nuclease domain-containing protein [Citrobacter]
MIPVIPPKRQDGKSSFGDLVSYVSVRDEPRDDDLMDAVKASGTRPEMPHSSRFSRLVDYATKLRDESFIS
LVDVMPDGGEWVNFYGVTCFHNCTSIETAAEEMEFTARQAKHSRSNSDPVFHYLLSWQSHESPRPEQVYD
SVRHSLKRLGMGDHQFVAAIHTDTDNLHVHVAVNRVHPGTGYLNWLSYSQEKLHKACRELELKHGFAPDN
GCYVIAPDNRIVRRTSVERDRQSAWTRGRNQSLKEYVADTAIASLRESPVTGWSSLHQRFAKEGLFLSAD
NGELKVKDGWNRERPGVTLSAFGHAFDTDKLIRKMGEFIPPAQDIFAQVPAVGRYEPDKIIVSSRPERMT
EKESLSDYAIARLRAPLIALDQDPSQRTIQSVHTLLAQSGLYLKEQHDRLVICDGYDPTRTPVRAERVWP
ALTKAVLNSYKGGWQPVPKDIFQQVPPAEQFTGGGLEPTPVSDREWRKLRTGSGPQGALKREIFSDKESL
WGYAISHCRQEIETLISSKHFSWERCHEQFAKQGLLLVREQQGLVVMDAYNREQTPVKASHVHPDLTLAR
AEPHAGSFVPVPADIFKRIKPVSRYNPELAVSDRDIPGMKRDPELRRQRREARAAAREDLKARYAAWRAN
WQRPDLHGRERYQVIHDECRIRKARIRIEHRDPLVRKLHYHIAELQRMQALITLKEAMKAERAELIGQGK
WYPPSYRQWVEAEAVKGDKAAISQLRGWDYRDRRGNAVKATTDSRCVVICEPGGTPIYSGVPGLQASLKK
NGRVQFRDPETGRHICTDYGDRVIFRNTGDYDALKRDMIKVAPVLFSRSPTVAMAPEGNDEQFNTAFAKM
VAWHNVNHRDEGEYQISRPDVDQVRVESEIRFSETMSQVSNSEPARNEPGSSWKPPSPL
MIPVIPPKRQDGKSSFGDLVSYVSVRDEPRDDDLMDAVKASGTRPEMPHSSRFSRLVDYATKLRDESFIS
LVDVMPDGGEWVNFYGVTCFHNCTSIETAAEEMEFTARQAKHSRSNSDPVFHYLLSWQSHESPRPEQVYD
SVRHSLKRLGMGDHQFVAAIHTDTDNLHVHVAVNRVHPGTGYLNWLSYSQEKLHKACRELELKHGFAPDN
GCYVIAPDNRIVRRTSVERDRQSAWTRGRNQSLKEYVADTAIASLRESPVTGWSSLHQRFAKEGLFLSAD
NGELKVKDGWNRERPGVTLSAFGHAFDTDKLIRKMGEFIPPAQDIFAQVPAVGRYEPDKIIVSSRPERMT
EKESLSDYAIARLRAPLIALDQDPSQRTIQSVHTLLAQSGLYLKEQHDRLVICDGYDPTRTPVRAERVWP
ALTKAVLNSYKGGWQPVPKDIFQQVPPAEQFTGGGLEPTPVSDREWRKLRTGSGPQGALKREIFSDKESL
WGYAISHCRQEIETLISSKHFSWERCHEQFAKQGLLLVREQQGLVVMDAYNREQTPVKASHVHPDLTLAR
AEPHAGSFVPVPADIFKRIKPVSRYNPELAVSDRDIPGMKRDPELRRQRREARAAAREDLKARYAAWRAN
WQRPDLHGRERYQVIHDECRIRKARIRIEHRDPLVRKLHYHIAELQRMQALITLKEAMKAERAELIGQGK
WYPPSYRQWVEAEAVKGDKAAISQLRGWDYRDRRGNAVKATTDSRCVVICEPGGTPIYSGVPGLQASLKK
NGRVQFRDPETGRHICTDYGDRVIFRNTGDYDALKRDMIKVAPVLFSRSPTVAMAPEGNDEQFNTAFAKM
VAWHNVNHRDEGEYQISRPDVDQVRVESEIRFSETMSQVSNSEPARNEPGSSWKPPSPL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 10718 | GenBank | WP_115490298 |
| Name | trbC_GHC22_RS23840_pH12-1 |
UniProt ID | _ |
| Length | 766 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 766 a.a. Molecular weight: 87341.52 Da Isoelectric Point: 6.7428
>WP_115490298.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Citrobacter]
MNNTPVNQALMKRNAWNSPVLELMRDHLLYAGIMAGSVVAGFIWPLAIPLFLLLAIVASISFGTHRWRMP
MRMPVHLNKVDPSEDRKVRRSFFRAFPSLFQYETTQESKGKGIFYIGYKRMNDIGRELWLTLDDLTRHVM
FFASTGGGKTETIYAWMLNSFCWGRGFTFVDGKAQNDTARTCYYLARRFGREDDVEYINFMNSGMSRSEM
IQNGDKSRPQSHQWNPFCYSTEAFVAETMQSMLPTNVQGGEWQSRAIAMNKALVFGTKFYCVRENKTMSL
QLLREFMPLEKLAGLYCRAVDDQWPEEAVSPLYNYLVDVPGFDMATVRQPSAWTEEPRKQHSYLTGQFLE
TFNTFTETFGNIFAEDAGDIDIRDSIHSDRILLVLIPAMNTSQHTTSALGRMFVTQQSMILARDLGHKLE
GLDSEALEITKYKGKFPYMNYLDEVGAYYTERIAVQATQVRSLEFALVMMSQDQERIENQTSAANVATLM
QNAGTKIAGKIVSDDKTAKTISNAAGQEARANMVSLQRQEGIIGTSWIDGDHINIQMENKINVQDLIKLQ
PGENYTVFQGDPVPGASFYIEPQDKTCNAPVIINRYITINPPTLKHLRRLVPRTAQRRLPAPENVSRIIG
VLTEKPSRRRKAARTEPWKIIDTFQQRLANRQEAHNLMTEYDMDHRGREENLWAEALEIISTTTMAERTI
RYITLNKPESESAEVNAEDIQLAPDAILRNLTLPQPASVPPARHRNEPLIPPKQTDSPQPDMEWPE
MNNTPVNQALMKRNAWNSPVLELMRDHLLYAGIMAGSVVAGFIWPLAIPLFLLLAIVASISFGTHRWRMP
MRMPVHLNKVDPSEDRKVRRSFFRAFPSLFQYETTQESKGKGIFYIGYKRMNDIGRELWLTLDDLTRHVM
FFASTGGGKTETIYAWMLNSFCWGRGFTFVDGKAQNDTARTCYYLARRFGREDDVEYINFMNSGMSRSEM
IQNGDKSRPQSHQWNPFCYSTEAFVAETMQSMLPTNVQGGEWQSRAIAMNKALVFGTKFYCVRENKTMSL
QLLREFMPLEKLAGLYCRAVDDQWPEEAVSPLYNYLVDVPGFDMATVRQPSAWTEEPRKQHSYLTGQFLE
TFNTFTETFGNIFAEDAGDIDIRDSIHSDRILLVLIPAMNTSQHTTSALGRMFVTQQSMILARDLGHKLE
GLDSEALEITKYKGKFPYMNYLDEVGAYYTERIAVQATQVRSLEFALVMMSQDQERIENQTSAANVATLM
QNAGTKIAGKIVSDDKTAKTISNAAGQEARANMVSLQRQEGIIGTSWIDGDHINIQMENKINVQDLIKLQ
PGENYTVFQGDPVPGASFYIEPQDKTCNAPVIINRYITINPPTLKHLRRLVPRTAQRRLPAPENVSRIIG
VLTEKPSRRRKAARTEPWKIIDTFQQRLANRQEAHNLMTEYDMDHRGREENLWAEALEIISTTTMAERTI
RYITLNKPESESAEVNAEDIQLAPDAILRNLTLPQPASVPPARHRNEPLIPPKQTDSPQPDMEWPE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 64379..101206
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| GHC22_RS23825 | 60901..61134 | + | 234 | WP_040231217 | hypothetical protein | - |
| GHC22_RS23830 | 61155..61670 | + | 516 | WP_167335593 | type II toxin-antitoxin system YafO family toxin | - |
| GHC22_RS23835 | 61733..62002 | - | 270 | WP_115490299 | hypothetical protein | - |
| GHC22_RS23840 | 62075..64375 | - | 2301 | WP_115490298 | F-type conjugative transfer protein TrbC | - |
| GHC22_RS23845 | 64379..65578 | - | 1200 | WP_153754028 | thioredoxin fold domain-containing protein | trbB |
| GHC22_RS23850 | 65631..66839 | - | 1209 | WP_153754029 | conjugal transfer protein TrbA | trbA |
| GHC22_RS23855 | 67150..67782 | - | 633 | WP_153754030 | plasmid IncI1-type surface exclusion protein ExcA | - |
| GHC22_RS23860 | 67844..70003 | - | 2160 | WP_153754031 | DotA/TraY family protein | traY |
| GHC22_RS23865 | 70061..70603 | - | 543 | WP_153754032 | conjugal transfer protein TraX | - |
| GHC22_RS23870 | 70600..71802 | - | 1203 | WP_153754033 | conjugal transfer protein TraW | traW |
| GHC22_RS23875 | 71815..72387 | - | 573 | WP_153754034 | conjugal transfer protein TraV | traV |
| GHC22_RS23880 | 72384..75437 | - | 3054 | WP_153754035 | ATP-binding protein | traU |
| GHC22_RS23885 | 75509..76240 | - | 732 | WP_228553287 | conjugal transfer protein TraT | traT |
| GHC22_RS23890 | 76424..76828 | - | 405 | WP_040231201 | DUF6750 family protein | traR |
| GHC22_RS23895 | 76862..77386 | - | 525 | WP_040231200 | conjugal transfer protein TraQ | traQ |
| GHC22_RS23900 | 77386..78159 | - | 774 | WP_153754036 | conjugal transfer protein TraP | traP |
| GHC22_RS23905 | 78140..79456 | - | 1317 | WP_153754037 | conjugal transfer protein TraO | traO |
| GHC22_RS23910 | 79456..80439 | - | 984 | WP_123924694 | DotH/IcmK family type IV secretion protein | traN |
| GHC22_RS23915 | 80450..81151 | - | 702 | WP_040231195 | DotI/IcmL/TraM family protein | traM |
| GHC22_RS23920 | 81148..81507 | - | 360 | WP_228553289 | conjugal transfer protein | traL |
| GHC22_RS24780 | 81525..83402 | - | 1878 | WP_228553301 | LPD7 domain-containing protein | - |
| GHC22_RS24785 | 84780..86012 | - | 1233 | Protein_90 | DUF5710 domain-containing protein | - |
| GHC22_RS23930 | 86374..86913 | - | 540 | WP_153754039 | hypothetical protein | - |
| GHC22_RS23935 | 86954..87241 | - | 288 | WP_040231189 | hypothetical protein | traK |
| GHC22_RS23940 | 87238..88398 | - | 1161 | WP_153754040 | plasmid transfer ATPase TraJ | virB11 |
| GHC22_RS23945 | 88410..89216 | - | 807 | WP_153754041 | type IV secretory system conjugative DNA transfer family protein | traI |
| GHC22_RS23950 | 89213..89686 | - | 474 | WP_040231184 | DotD/TraH family lipoprotein | - |
| GHC22_RS23955 | 89819..91027 | - | 1209 | WP_153754042 | conjugal transfer protein TraF | - |
| GHC22_RS23960 | 91130..91966 | - | 837 | WP_040231179 | hypothetical protein | traE |
| GHC22_RS23965 | 92111..92587 | - | 477 | WP_123924625 | hypothetical protein | - |
| GHC22_RS23970 | 92607..93767 | - | 1161 | WP_228553303 | site-specific integrase | - |
| GHC22_RS23975 | 94352..94648 | + | 297 | WP_153754043 | hypothetical protein | - |
| GHC22_RS23980 | 95486..96709 | - | 1224 | WP_153754044 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| GHC22_RS23985 | 96706..97380 | - | 675 | WP_040231341 | prepilin peptidase | - |
| GHC22_RS23990 | 97365..97847 | - | 483 | WP_123924696 | lytic transglycosylase domain-containing protein | virB1 |
| GHC22_RS23995 | 97932..98546 | - | 615 | WP_052463753 | type 4 pilus major pilin | - |
| GHC22_RS24000 | 98560..99654 | - | 1095 | WP_040231337 | type II secretion system F family protein | - |
| GHC22_RS24005 | 99647..101206 | - | 1560 | WP_153754045 | GspE/PulE family protein | virB11 |
| GHC22_RS24010 | 101209..101709 | - | 501 | WP_153754046 | type IV pilus biogenesis protein PilP | - |
| GHC22_RS24015 | 101693..102997 | - | 1305 | WP_153754047 | type 4b pilus protein PilO2 | - |
| GHC22_RS24020 | 102999..104675 | - | 1677 | WP_040231333 | PilN family type IVB pilus formation outer membrane protein | - |
| GHC22_RS24025 | 104690..105100 | - | 411 | WP_228553291 | type IV pilus biogenesis protein PilM | - |
Host bacterium
| ID | 14863 | GenBank | NZ_CP045838 |
| Plasmid name | pH12-1 | Incompatibility group | - |
| Plasmid size | 110138 bp | Coordinate of oriT [Strand] | 57652..57736 [-] |
| Host baterium | Citrobacter sp. H12-3-2 |
Cargo genes
| Drug resistance gene | blaCTX-M-14, fosA3 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |