Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114419 |
| Name | oriT_pN03G-1 |
| Organism | Bradyrhizobium japonicum strain N03G-Bj |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP126011 (146975..147016 [+], 42 nt) |
| oriT length | 42 nt |
| IRs (inverted repeats) | 9..15, 20..26 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 42 nt
>oriT_pN03G-1
AAAGCAGGACGTCGCGGATGCGACGTATTACTGCGCCCTTGG
AAAGCAGGACGTCGCGGATGCGACGTATTACTGCGCCCTTGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file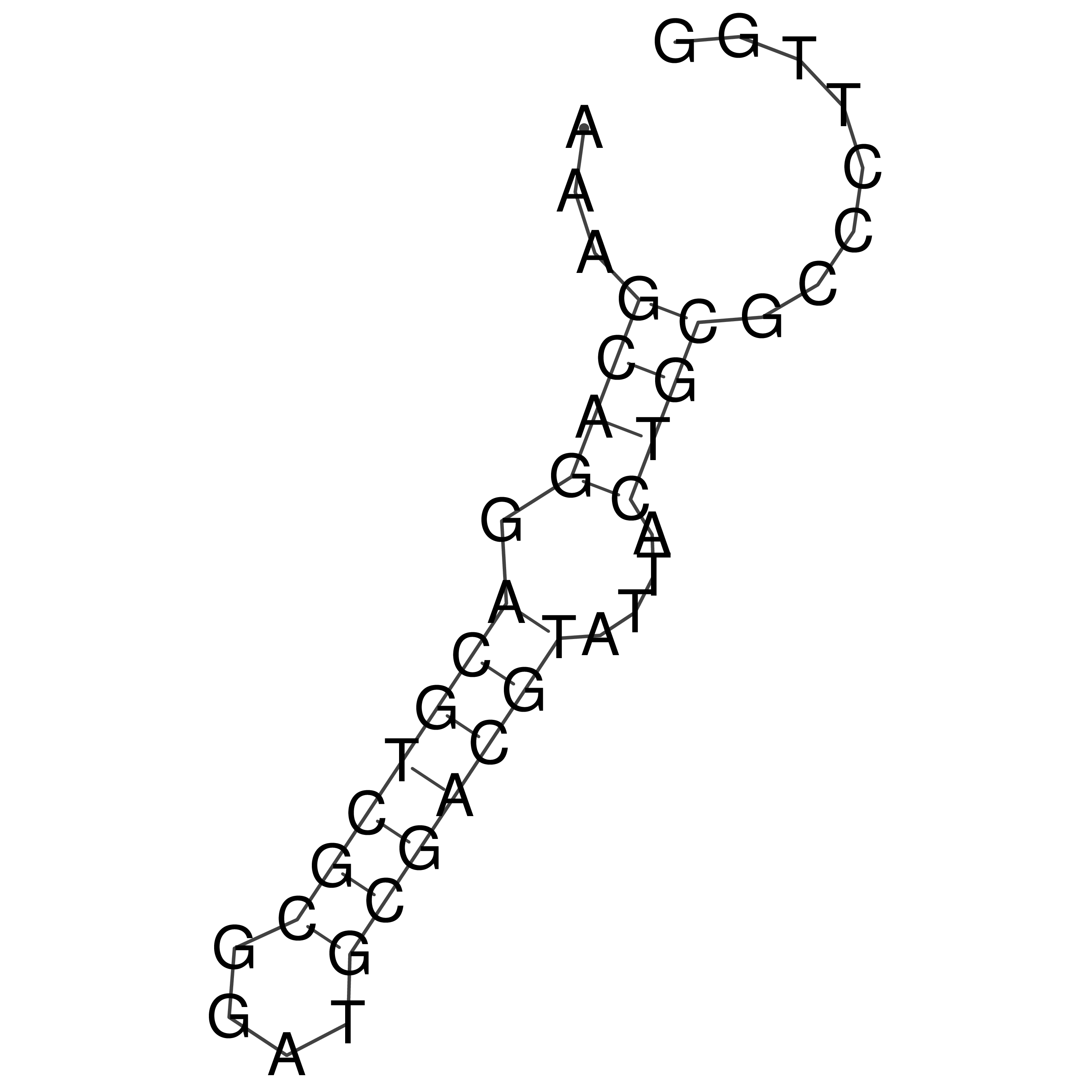
Relaxase
| ID | 9239 | GenBank | WP_212479306 |
| Name | traA_QIH85_RS47710_pN03G-1 |
UniProt ID | _ |
| Length | 1102 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1102 a.a. Molecular weight: 121918.26 Da Isoelectric Point: 9.9984
>WP_212479306.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Bradyrhizobium]
MAITHFTPQLISRGSGRSAVLSAAYRHCARMEHQAEGRTVDYSAKRGLRYEEFLLPPDAPQWARELIADR
SVAGAAEAFWNAIEAFEKRSDAQLAKEFIIALPVELTIDQNVALVREFVATQVLARGQVADWVFHDEPGN
PHIHLMTTLRPLGEDGFGPKKVPVIGHDGAPLRNAAGKIVYRLWSGEKAEFLAQREGWLDLQNRHLALAG
LDIRVDGRSYAERGVDLAPTTHIGVATKAIDRKGKKAGWSPKLERVALHEERKTDNRQRILRRPEIVLDL
VSREMSVFTERDIARVLHRYVDDAGTFQHLMARILQNPEALRLEGECIALATGIREPAKYTTRELIRLEA
EMAHRAVWLAGRSSHGVREKVLAAAFERHARLSEEQRVAIAHVASVGRIAAVVGRAGAGKTTMMKAAREV
WEAAGYRVVGAALAGKAAEGLEKEAGIASRTLASWELAWSNGREQLDDKTIFVLDEAGMVSSRQMALFVE
AVVKTGAKLALVGDPDQLQPIEAGAAFRAIAERTGYAELETIYRQREQWMRDASLDLARGRVAEALQAYG
ANGKLEARKYKAQAVEALIEDWNWDYDPAKSTLILAHLRRDVRVLNEMARAKLIERGIVEDGHAFRTEDG
PRSFATGDQIVFLKNEGSLGVKNGMIARVIEARPGWFAAMIGEGEARRRVEVDQRFYANVDHGYATTIHK
SQGATVDRVKVLATLSLDRHLAYVALTRHREDVALYYGAISFGKAGGLTKILSQSRAKETTLDYERGPSY
RAALRFAEARGLHLMRVARTLVADKVRWTVRQKQKLADLGARLLALGARLGLGAARKQTIPNGAKEAKPM
VAGITTFAKSIDQVVEDKLAADPGLKKQWEEVSTRFRFVFAEPESAFRKVDVDSMLKDKATAQSTLSKLA
EKPESFGALKGKTGLLASRADKQDRQRADLNVPALARDLERFLRLRSEAEHRYEAEERAARLKVAVDIPA
LSPSAKQTLERIRDAIDRNDLPAALEFALEDKMIEAELEGFARAVSERFGERSFVGLAAKDTDGKTFETL
SGGMNAAQKEGLKAAWPDLRTAQQLAAQQRTAVALKEAKTLRQTQSKGLSLQ
MAITHFTPQLISRGSGRSAVLSAAYRHCARMEHQAEGRTVDYSAKRGLRYEEFLLPPDAPQWARELIADR
SVAGAAEAFWNAIEAFEKRSDAQLAKEFIIALPVELTIDQNVALVREFVATQVLARGQVADWVFHDEPGN
PHIHLMTTLRPLGEDGFGPKKVPVIGHDGAPLRNAAGKIVYRLWSGEKAEFLAQREGWLDLQNRHLALAG
LDIRVDGRSYAERGVDLAPTTHIGVATKAIDRKGKKAGWSPKLERVALHEERKTDNRQRILRRPEIVLDL
VSREMSVFTERDIARVLHRYVDDAGTFQHLMARILQNPEALRLEGECIALATGIREPAKYTTRELIRLEA
EMAHRAVWLAGRSSHGVREKVLAAAFERHARLSEEQRVAIAHVASVGRIAAVVGRAGAGKTTMMKAAREV
WEAAGYRVVGAALAGKAAEGLEKEAGIASRTLASWELAWSNGREQLDDKTIFVLDEAGMVSSRQMALFVE
AVVKTGAKLALVGDPDQLQPIEAGAAFRAIAERTGYAELETIYRQREQWMRDASLDLARGRVAEALQAYG
ANGKLEARKYKAQAVEALIEDWNWDYDPAKSTLILAHLRRDVRVLNEMARAKLIERGIVEDGHAFRTEDG
PRSFATGDQIVFLKNEGSLGVKNGMIARVIEARPGWFAAMIGEGEARRRVEVDQRFYANVDHGYATTIHK
SQGATVDRVKVLATLSLDRHLAYVALTRHREDVALYYGAISFGKAGGLTKILSQSRAKETTLDYERGPSY
RAALRFAEARGLHLMRVARTLVADKVRWTVRQKQKLADLGARLLALGARLGLGAARKQTIPNGAKEAKPM
VAGITTFAKSIDQVVEDKLAADPGLKKQWEEVSTRFRFVFAEPESAFRKVDVDSMLKDKATAQSTLSKLA
EKPESFGALKGKTGLLASRADKQDRQRADLNVPALARDLERFLRLRSEAEHRYEAEERAARLKVAVDIPA
LSPSAKQTLERIRDAIDRNDLPAALEFALEDKMIEAELEGFARAVSERFGERSFVGLAAKDTDGKTFETL
SGGMNAAQKEGLKAAWPDLRTAQQLAAQQRTAVALKEAKTLRQTQSKGLSLQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 10700 | GenBank | WP_028152596 |
| Name | t4cp2_QIH85_RS47635_pN03G-1 |
UniProt ID | _ |
| Length | 817 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 817 a.a. Molecular weight: 91386.01 Da Isoelectric Point: 6.3078
>WP_028152596.1 MULTISPECIES: conjugal transfer protein TrbE [Bradyrhizobium]
MVALRSFRHSGPSFADLVPYAGLVDDGIVLLKDGSLMAGWYFAGPDSESSTDAERNDVSRQINAILARLG
SGWMIQVEALRIPTTDYPARKDGHFPDAVTRAIDEERRGHFERERGHYESRHALILTWRPPERRKSGLAK
YVYSDKDSRSASFADTALGNFRTSIREIEQYLSNLLSIQRMRTRETAERGDARIARYDELFQFVRFCITG
ENHPVRLPEVPMYLDWLVTAEYQHGLTPTVDGRYVGVVAIDGLPAESWPGILNILDLMPLTYRWSSRFMF
LDEIEARERLERTRKKWQQKVRPFFDQLFQTQSRSIDQDAMAMVAETQDAIAEAASQLVAYGYYTPVIVL
QDHDAERLREKCEGIRRVVQAEGFGARIETLNATEAFLGSLPGNWYANIREPLIHTRNLADLLPLNSVWS
GSPVAPCPFYPEGSPPLMQVASGSTPFRLNLHTDDVGHSLIFGPTGSGKSTLLALIAAQFRRYRDAQIFA
FDKGRSLMPLTLAAGGDHYEIGGGAGEGPQLAFCPLAELATDADRAWASEWIETLVVLQGVSVTPDHRNA
ISRQVALMAAAPGRSLADFVSGVQMREIKDALHHYTVDGPMGHLLDAERDGLVLGGFQTFEIEELMNMGE
RSLVPVLTYLFRRIEKRLTGAPSLILLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAERS
GIIDVLKESCPTKICLPNGAAREPGTREFYERLGFNERQIEIVSNAIPKREYYVTSPAGRRLFDMALGPV
ALAFVGASGRDDLKRITELKAVHGRTWPANWLQTRGIAHAETLLADD
MVALRSFRHSGPSFADLVPYAGLVDDGIVLLKDGSLMAGWYFAGPDSESSTDAERNDVSRQINAILARLG
SGWMIQVEALRIPTTDYPARKDGHFPDAVTRAIDEERRGHFERERGHYESRHALILTWRPPERRKSGLAK
YVYSDKDSRSASFADTALGNFRTSIREIEQYLSNLLSIQRMRTRETAERGDARIARYDELFQFVRFCITG
ENHPVRLPEVPMYLDWLVTAEYQHGLTPTVDGRYVGVVAIDGLPAESWPGILNILDLMPLTYRWSSRFMF
LDEIEARERLERTRKKWQQKVRPFFDQLFQTQSRSIDQDAMAMVAETQDAIAEAASQLVAYGYYTPVIVL
QDHDAERLREKCEGIRRVVQAEGFGARIETLNATEAFLGSLPGNWYANIREPLIHTRNLADLLPLNSVWS
GSPVAPCPFYPEGSPPLMQVASGSTPFRLNLHTDDVGHSLIFGPTGSGKSTLLALIAAQFRRYRDAQIFA
FDKGRSLMPLTLAAGGDHYEIGGGAGEGPQLAFCPLAELATDADRAWASEWIETLVVLQGVSVTPDHRNA
ISRQVALMAAAPGRSLADFVSGVQMREIKDALHHYTVDGPMGHLLDAERDGLVLGGFQTFEIEELMNMGE
RSLVPVLTYLFRRIEKRLTGAPSLILLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAERS
GIIDVLKESCPTKICLPNGAAREPGTREFYERLGFNERQIEIVSNAIPKREYYVTSPAGRRLFDMALGPV
ALAFVGASGRDDLKRITELKAVHGRTWPANWLQTRGIAHAETLLADD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 10701 | GenBank | WP_028152606 |
| Name | traG_QIH85_RS47725_pN03G-1 |
UniProt ID | _ |
| Length | 640 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 640 a.a. Molecular weight: 68584.77 Da Isoelectric Point: 9.2825
>WP_028152606.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Bradyrhizobium]
MTASRLLLLIVPVALMLGTVIGLTGIEHSLAGLGKTEAQRLLVARAGIAMPYIVAAAIGVVFLFAAAGSA
NIRTAAWSVVAGCAAAISMAAAREAIRLSAILANAVHGRSPPVQIDPAIMVGAGIALMAGIFALRVAVKG
NAAFAAPMPKRVRGRRALHGDADWMLVPEAAGLFSDAGGIVVGERYRVDKDSAAAEPFRAMDRQSWGIGG
RAPLLCFDATFGSSHGIVFAGSGGFKTTSVTIPTALKWGGALVVLDPSNEVAPMVIEHRRAAGRTVHVLD
PREPGTGFNALDWIGRFGGTKEEDIAAVASWIMSDSGGVRGVRDDFFRASALQLVTALIADVCLSGHTED
KNQTLRQVRANLSEPEPKLRARLQAIYDNSESQFVKENVAVFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGSTFVSDDLAGGATDVFINLDLKTLETHAGLARVIIGAFMNAIYNRNGEVTGRALFLLDEVARLGF
MRILETARDAGRKYGITLLLLFQSIGQMRETYGGRDAASKWFESASWISFAAVNDPETAEYISKRCGMTT
VEIDQVSRSSQASGSSRTRSKQLASRPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMRACVG
KNRFQPEEKT
MTASRLLLLIVPVALMLGTVIGLTGIEHSLAGLGKTEAQRLLVARAGIAMPYIVAAAIGVVFLFAAAGSA
NIRTAAWSVVAGCAAAISMAAAREAIRLSAILANAVHGRSPPVQIDPAIMVGAGIALMAGIFALRVAVKG
NAAFAAPMPKRVRGRRALHGDADWMLVPEAAGLFSDAGGIVVGERYRVDKDSAAAEPFRAMDRQSWGIGG
RAPLLCFDATFGSSHGIVFAGSGGFKTTSVTIPTALKWGGALVVLDPSNEVAPMVIEHRRAAGRTVHVLD
PREPGTGFNALDWIGRFGGTKEEDIAAVASWIMSDSGGVRGVRDDFFRASALQLVTALIADVCLSGHTED
KNQTLRQVRANLSEPEPKLRARLQAIYDNSESQFVKENVAVFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGSTFVSDDLAGGATDVFINLDLKTLETHAGLARVIIGAFMNAIYNRNGEVTGRALFLLDEVARLGF
MRILETARDAGRKYGITLLLLFQSIGQMRETYGGRDAASKWFESASWISFAAVNDPETAEYISKRCGMTT
VEIDQVSRSSQASGSSRTRSKQLASRPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMRACVG
KNRFQPEEKT
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 129196..138772
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| QIH85_RS47600 (QIH85_47600) | 124423..125647 | - | 1225 | Protein_124 | plasmid replication protein RepC | - |
| QIH85_RS47605 (QIH85_47605) | 125874..126887 | - | 1014 | WP_049808001 | plasmid partitioning protein RepB | - |
| QIH85_RS47610 (QIH85_47610) | 126884..128077 | - | 1194 | WP_028152592 | plasmid partitioning protein RepA | - |
| QIH85_RS47615 (QIH85_47615) | 128570..129199 | + | 630 | WP_063629878 | acyl-homoserine-lactone synthase | - |
| QIH85_RS47620 (QIH85_47620) | 129196..130164 | + | 969 | WP_060913174 | P-type conjugative transfer ATPase TrbB | virB11 |
| QIH85_RS47625 (QIH85_47625) | 130154..130543 | + | 390 | WP_028152594 | TrbC/VirB2 family protein | virB2 |
| QIH85_RS47630 (QIH85_47630) | 130536..130835 | + | 300 | WP_028152595 | conjugal transfer protein TrbD | virB3 |
| QIH85_RS47635 (QIH85_47635) | 130845..133298 | + | 2454 | WP_028152596 | conjugal transfer protein TrbE | virb4 |
| QIH85_RS47640 (QIH85_47640) | 133270..134088 | + | 819 | WP_038953896 | P-type conjugative transfer protein TrbJ | virB5 |
| QIH85_RS47645 (QIH85_47645) | 134082..134294 | + | 213 | WP_028152598 | entry exclusion protein TrbK | - |
| QIH85_RS47650 (QIH85_47650) | 134288..135475 | + | 1188 | WP_038945592 | P-type conjugative transfer protein TrbL | virB6 |
| QIH85_RS47655 (QIH85_47655) | 135490..136152 | + | 663 | WP_028152600 | conjugal transfer protein TrbF | virB8 |
| QIH85_RS47660 (QIH85_47660) | 136170..136994 | + | 825 | WP_028152601 | P-type conjugative transfer protein TrbG | virB9 |
| QIH85_RS47665 (QIH85_47665) | 136999..137463 | + | 465 | WP_028152602 | conjugal transfer protein TrbH | - |
| QIH85_RS47670 (QIH85_47670) | 137474..138772 | + | 1299 | WP_028152603 | IncP-type conjugal transfer protein TrbI | virB10 |
| QIH85_RS47675 (QIH85_47675) | 139047..139958 | + | 912 | WP_232674644 | autoinducer binding domain-containing protein | - |
| QIH85_RS47680 (QIH85_47680) | 139967..140836 | - | 870 | WP_232674645 | transcriptional repressor TraM | - |
| QIH85_RS49635 | 140841..141089 | - | 249 | WP_080586789 | DUF2167 domain-containing protein | - |
| QIH85_RS47685 (QIH85_47685) | 141098..141688 | - | 591 | WP_049807966 | DUF2167 domain-containing protein | - |
| QIH85_RS47690 (QIH85_47690) | 141827..141946 | + | 120 | Protein_143 | type II toxin-antitoxin system VapC family toxin | - |
| QIH85_RS47695 (QIH85_47695) | 141977..142582 | - | 606 | WP_035680223 | TraH family protein | - |
| QIH85_RS47700 (QIH85_47700) | 142597..142919 | - | 323 | Protein_145 | nitrilase-related carbon-nitrogen hydrolase | - |
| QIH85_RS47705 (QIH85_47705) | 142977..143480 | - | 504 | WP_038953892 | conjugative transfer signal peptidase TraF | - |
Host bacterium
| ID | 14854 | GenBank | NZ_CP126011 |
| Plasmid name | pN03G-1 | Incompatibility group | - |
| Plasmid size | 165114 bp | Coordinate of oriT [Strand] | 146975..147016 [+] |
| Host baterium | Bradyrhizobium japonicum strain N03G-Bj |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | bscN |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |