Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114352 |
| Name | oriT_pRlBT01a |
| Organism | Rhizobium leguminosarum strain BT01 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP125636 (462770..462798 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pRlBT01a
AGGGCGCAATATACGTCGCTGTCGCGACG
AGGGCGCAATATACGTCGCTGTCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file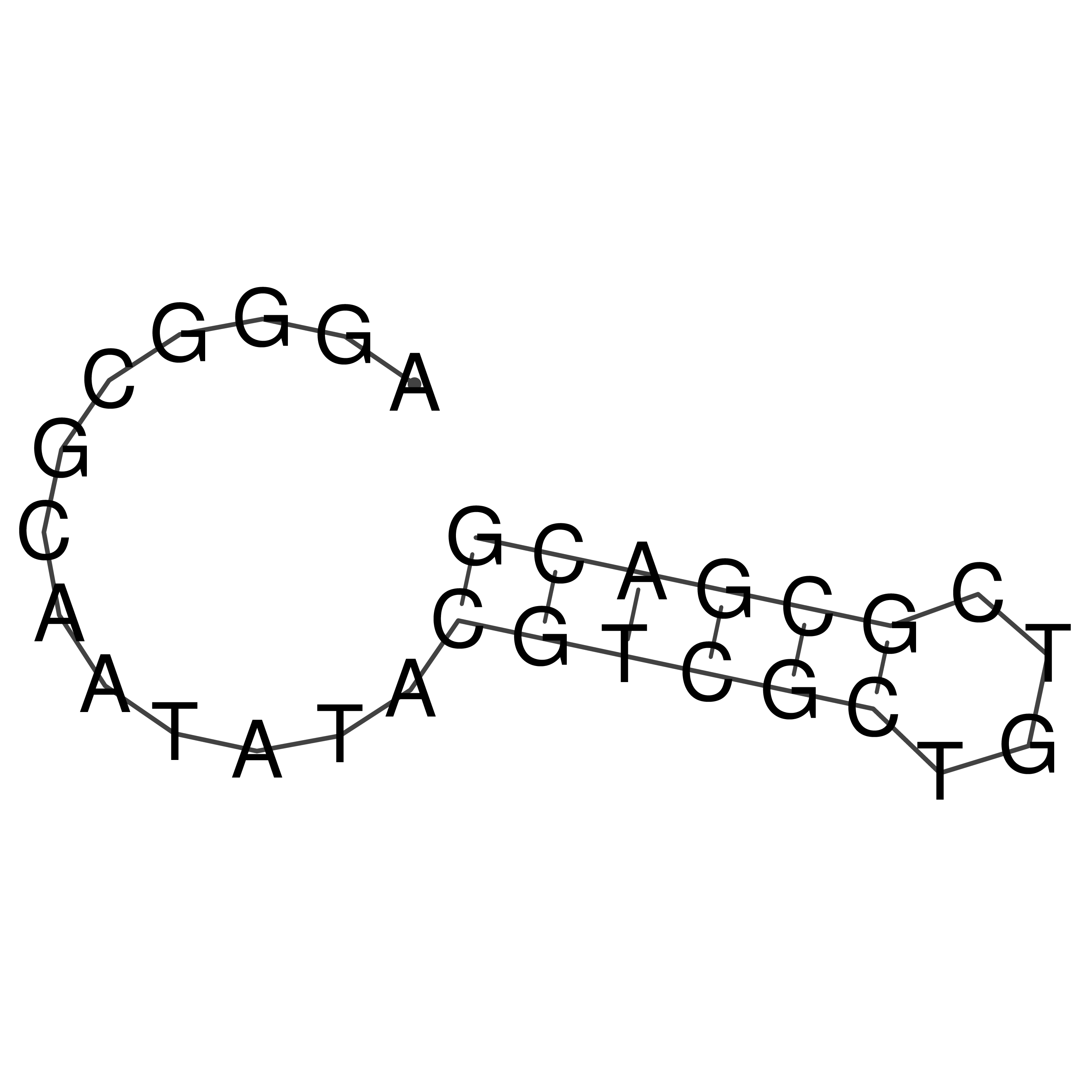
Relaxase
| ID | 9200 | GenBank | WP_077991369 |
| Name | traA_QMO81_RS26440_pRlBT01a |
UniProt ID | _ |
| Length | 1565 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1565 a.a. Molecular weight: 172927.47 Da Isoelectric Point: 6.8704
>WP_077991369.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAIMFVRAQVISRGSGRSIVSAAAYRHRARMMDEQAGTSFSYRGGASELMHEELALPGQIPTWLREAIAG
QLVAKASEALWNAVDAFETRADAQLARELIIALPEELTRAENITLVREFIRDNLISKGMIADWVYHDKEG
NPHIHLMTTLRPLTEEGFGAKKVPVLGEDGKPLRVVTPDRPNGKIVYKVWAGDKETMKAWKIAWAETANR
HLALAGHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGRELHFAPADLARRQEMADRLLSEPELLLKQ
LGNERSTFDERDIARALHRYVDDPTDFANIRARLMASDQLVILKPQEIEAETGKVSEPAMFTTREMLRIE
YDMAQSARVLSERRGFGVSERIVTVAIERVESIESGDPKNPFRLDAEQVDAVRHVTGDGDIAAIVGLAGA
GKSTLLAAARLAWESEGHRVIGAALAGKAAEGLQDSSGIKSRTLASWELAWANGRDTLHRGDVLVIDEAG
MVASQQMARVLKIAEEAEVKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQREAWARNASRLFA
RGEVEKGLDAYARHGHLVEAGSREETIDRIVSDWAAARREAIERSTSEGGDGRLRGDELLVLAHTNDDVR
KLNEALRSVMTQEGALSESRSFRSERGVREFAAGDRIIFLENARFLEPRAKHSGPQYVKNGMLGTVVSTG
DKRGDPLLSVLLDNGRKLVFSEDSYRHVDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRDR
VDLYAAKEDFAAKPEWGRKPRVDHAAGVTGELVETGEAKFRPDDEDADDSPYADVRADGGTVYRLWGVSL
PKALEEAGIQEGDTVTLRKDGVERVKVQITVVDEETGHKRYEEREVDRNVWTASQVETASARQERIERES
HRPELFNPLVERLSRSGAKTTTLDFESEASYRAHANDFARRRGLDHLSLAAAEMEESLTRRWAWIAEKRE
QVEKLWERASVALGFAIERERRVAYNEERSQTMVEATSSDARTASHYVSGASADETPYLIPPATSFVSSV
EEDARLAQLASPAWTEREVILRPLLQKIYRDPDAALISLNALASDIGVAPRRLADDLAAAPGRLGRLRGS
ELIVDGRAAREERNVAVAAVKELLPMARAHATEFRRNAERFELREQTRRAHMSLSIPALSERAMARLLEI
EAVRSQGGDDAYKTAFALAAKDRSVVQEIKAVSEALTARFGWSAFSAKADVIAERNIVERMPEDLTDEWR
GKLTRLFEAVRRFADEQHLAERRDRSKIVAGASVDLRTETEKGVGKENVPIPPMLAAVTEFKVPVDDEAR
SRALASPLYRQQRAALANAATTIWRDPAEVVGKIEELLQKGFAAERIGAAVTNNPAAYGALRGSDRLMDR
MLASGRERKEAVAAVPEAAARLRALGAAHLNALDAERQAITDERRRMAVAIPALSKAAEEALAHLTVEVS
KDSRKLSVSAASLDPGIGREFAAVSRALDERFGRNALVRGDKAIANVVPPTQRGAFAAMQERLKVLQQTV
RLQSSEQIIVERRQRAASRARGINL
MAIMFVRAQVISRGSGRSIVSAAAYRHRARMMDEQAGTSFSYRGGASELMHEELALPGQIPTWLREAIAG
QLVAKASEALWNAVDAFETRADAQLARELIIALPEELTRAENITLVREFIRDNLISKGMIADWVYHDKEG
NPHIHLMTTLRPLTEEGFGAKKVPVLGEDGKPLRVVTPDRPNGKIVYKVWAGDKETMKAWKIAWAETANR
HLALAGHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGRELHFAPADLARRQEMADRLLSEPELLLKQ
LGNERSTFDERDIARALHRYVDDPTDFANIRARLMASDQLVILKPQEIEAETGKVSEPAMFTTREMLRIE
YDMAQSARVLSERRGFGVSERIVTVAIERVESIESGDPKNPFRLDAEQVDAVRHVTGDGDIAAIVGLAGA
GKSTLLAAARLAWESEGHRVIGAALAGKAAEGLQDSSGIKSRTLASWELAWANGRDTLHRGDVLVIDEAG
MVASQQMARVLKIAEEAEVKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQREAWARNASRLFA
RGEVEKGLDAYARHGHLVEAGSREETIDRIVSDWAAARREAIERSTSEGGDGRLRGDELLVLAHTNDDVR
KLNEALRSVMTQEGALSESRSFRSERGVREFAAGDRIIFLENARFLEPRAKHSGPQYVKNGMLGTVVSTG
DKRGDPLLSVLLDNGRKLVFSEDSYRHVDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRDR
VDLYAAKEDFAAKPEWGRKPRVDHAAGVTGELVETGEAKFRPDDEDADDSPYADVRADGGTVYRLWGVSL
PKALEEAGIQEGDTVTLRKDGVERVKVQITVVDEETGHKRYEEREVDRNVWTASQVETASARQERIERES
HRPELFNPLVERLSRSGAKTTTLDFESEASYRAHANDFARRRGLDHLSLAAAEMEESLTRRWAWIAEKRE
QVEKLWERASVALGFAIERERRVAYNEERSQTMVEATSSDARTASHYVSGASADETPYLIPPATSFVSSV
EEDARLAQLASPAWTEREVILRPLLQKIYRDPDAALISLNALASDIGVAPRRLADDLAAAPGRLGRLRGS
ELIVDGRAAREERNVAVAAVKELLPMARAHATEFRRNAERFELREQTRRAHMSLSIPALSERAMARLLEI
EAVRSQGGDDAYKTAFALAAKDRSVVQEIKAVSEALTARFGWSAFSAKADVIAERNIVERMPEDLTDEWR
GKLTRLFEAVRRFADEQHLAERRDRSKIVAGASVDLRTETEKGVGKENVPIPPMLAAVTEFKVPVDDEAR
SRALASPLYRQQRAALANAATTIWRDPAEVVGKIEELLQKGFAAERIGAAVTNNPAAYGALRGSDRLMDR
MLASGRERKEAVAAVPEAAARLRALGAAHLNALDAERQAITDERRRMAVAIPALSKAAEEALAHLTVEVS
KDSRKLSVSAASLDPGIGREFAAVSRALDERFGRNALVRGDKAIANVVPPTQRGAFAAMQERLKVLQQTV
RLQSSEQIIVERRQRAASRARGINL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 10664 | GenBank | WP_077991366 |
| Name | traG_QMO81_RS24175_pRlBT01a |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70297.10 Da Isoelectric Point: 9.0817
>WP_077991366.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTPLRKPRPSLLLVLAPIAGTAFGAYVVGWRWQELAAVMSGKTEYWFLRFAPLPALLFGPLAGLLTVLAL
PLHRRRPVAIVSLVSFLAVAGFYALREYGRLAPSVESGAVSWSRALSYLDLVAVIGALMGFMAVAVAARI
STVVSEPVTRARRGIFGDAGWLSMSAAAKLFPPDGEIVVGERYRVDQEIVHELPFDPNDPATWGQGGKAS
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYSGPLICLDPSTEVAPMVAEHRAGALKREVMVLDPT
NPIMGFNVLDGIEASKQKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYEGRRS
LRSLRQIVSEPEPSVLAMLRDIQEHSGSAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNISASILRSYPGIARVIIGSLINAMVQADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNIGWDTKNNASRRSENVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDEAAK
VNRFVKPVL
MTPLRKPRPSLLLVLAPIAGTAFGAYVVGWRWQELAAVMSGKTEYWFLRFAPLPALLFGPLAGLLTVLAL
PLHRRRPVAIVSLVSFLAVAGFYALREYGRLAPSVESGAVSWSRALSYLDLVAVIGALMGFMAVAVAARI
STVVSEPVTRARRGIFGDAGWLSMSAAAKLFPPDGEIVVGERYRVDQEIVHELPFDPNDPATWGQGGKAS
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYSGPLICLDPSTEVAPMVAEHRAGALKREVMVLDPT
NPIMGFNVLDGIEASKQKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYEGRRS
LRSLRQIVSEPEPSVLAMLRDIQEHSGSAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNISASILRSYPGIARVIIGSLINAMVQADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNIGWDTKNNASRRSENVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDEAAK
VNRFVKPVL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 9338..18939
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| QMO81_RS24205 (QMO81_004846) | 5171..5567 | - | 397 | Protein_7 | type II toxin-antitoxin system VapC family toxin | - |
| QMO81_RS24210 (QMO81_004847) | 5564..5830 | - | 267 | WP_004676093 | type II toxin-antitoxin system Phd/YefM family antitoxin | - |
| QMO81_RS24215 (QMO81_004848) | 5915..6037 | - | 123 | WP_258719717 | hypothetical protein | - |
| QMO81_RS24220 (QMO81_004849) | 6217..6476 | - | 260 | Protein_10 | toprim domain-containing protein | - |
| QMO81_RS24225 (QMO81_004850) | 6787..7977 | + | 1191 | WP_077991364 | DUF1173 domain-containing protein | - |
| QMO81_RS24230 (QMO81_004851) | 8281..8727 | + | 447 | WP_077991362 | excisionase | - |
| QMO81_RS24235 (QMO81_004852) | 8717..9157 | + | 441 | WP_245304432 | PIN domain-containing protein | - |
| QMO81_RS24240 (QMO81_004853) | 9338..10366 | - | 1029 | WP_077991360 | P-type DNA transfer ATPase VirB11 | virB11 |
| QMO81_RS24245 (QMO81_004854) | 10374..11549 | - | 1176 | WP_077991358 | type IV secretion system protein VirB10 | virB10 |
| QMO81_RS24250 (QMO81_004855) | 11558..12415 | - | 858 | WP_077991357 | P-type conjugative transfer protein VirB9 | virB9 |
| QMO81_RS24255 (QMO81_004856) | 12412..13083 | - | 672 | WP_077991356 | virB8 family protein | virB8 |
| QMO81_RS24260 (QMO81_004857) | 13085..13369 | - | 285 | WP_077991355 | hypothetical protein | - |
| QMO81_RS24265 (QMO81_004858) | 13409..14341 | - | 933 | WP_077991353 | type IV secretion system protein | virB6 |
| QMO81_RS24270 (QMO81_004859) | 14344..14577 | - | 234 | WP_007636178 | EexN family lipoprotein | - |
| QMO81_RS24275 (QMO81_004860) | 14574..15275 | - | 702 | WP_077991351 | P-type DNA transfer protein VirB5 | virB5 |
| QMO81_RS24280 (QMO81_004861) | 15272..17638 | - | 2367 | WP_077991348 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| QMO81_RS24285 (QMO81_004862) | 17631..17969 | - | 339 | WP_077991347 | type IV secretion system protein VirB3 | virB3 |
| QMO81_RS24290 (QMO81_004863) | 17975..18274 | - | 300 | WP_077991346 | TrbC/VirB2 family protein | virB2 |
| QMO81_RS24295 (QMO81_004864) | 18271..18939 | - | 669 | WP_077991344 | transglycosylase SLT domain-containing protein | virB1 |
| QMO81_RS24300 (QMO81_004865) | 18943..19479 | - | 537 | WP_077991342 | hypothetical protein | - |
| QMO81_RS24305 (QMO81_004866) | 19578..19949 | + | 372 | WP_004673403 | hypothetical protein | - |
| QMO81_RS24310 (QMO81_004867) | 20718..20900 | + | 183 | WP_016737443 | hypothetical protein | - |
| QMO81_RS24315 (QMO81_004868) | 21011..21223 | - | 213 | Protein_29 | zincin-like metallopeptidase domain-containing protein | - |
| QMO81_RS24320 (QMO81_004869) | 21256..21807 | - | 552 | WP_077991339 | hypothetical protein | - |
| QMO81_RS24325 (QMO81_004870) | 21800..22381 | - | 582 | WP_077991337 | hypothetical protein | - |
Host bacterium
| ID | 14787 | GenBank | NZ_CP125636 |
| Plasmid name | pRlBT01a | Incompatibility group | - |
| Plasmid size | 463472 bp | Coordinate of oriT [Strand] | 462770..462798 [-] |
| Host baterium | Rhizobium leguminosarum strain BT01 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | hsiC1/vipB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nodD, nodA, nifH, nifD, nifK, nifX, mLTONO_5203, fixN, fixO, nodC, nodS, nodI, nodJ, nifT, nifZ, nifB, nifA, fixX, fixC, fixB, fixA, nifW, nifS, nifE, nifN |
| Anti-CRISPR | - |