Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114294 |
| Name | oriT_p12-6919.1 |
| Organism | Salmonella enterica subsp. enterica serovar 14[5]12:i:- strain PNCS014868 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP039604 (8443..8531 [+], 89 nt) |
| oriT length | 89 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 89 nt
>oriT_p12-6919.1
GGGGTGTCGGGGCGAAGCCCTGACCAGATGGCAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTGCACATCCTGTCCCGTTTTTC
GGGGTGTCGGGGCGAAGCCCTGACCAGATGGCAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTGCACATCCTGTCCCGTTTTTC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file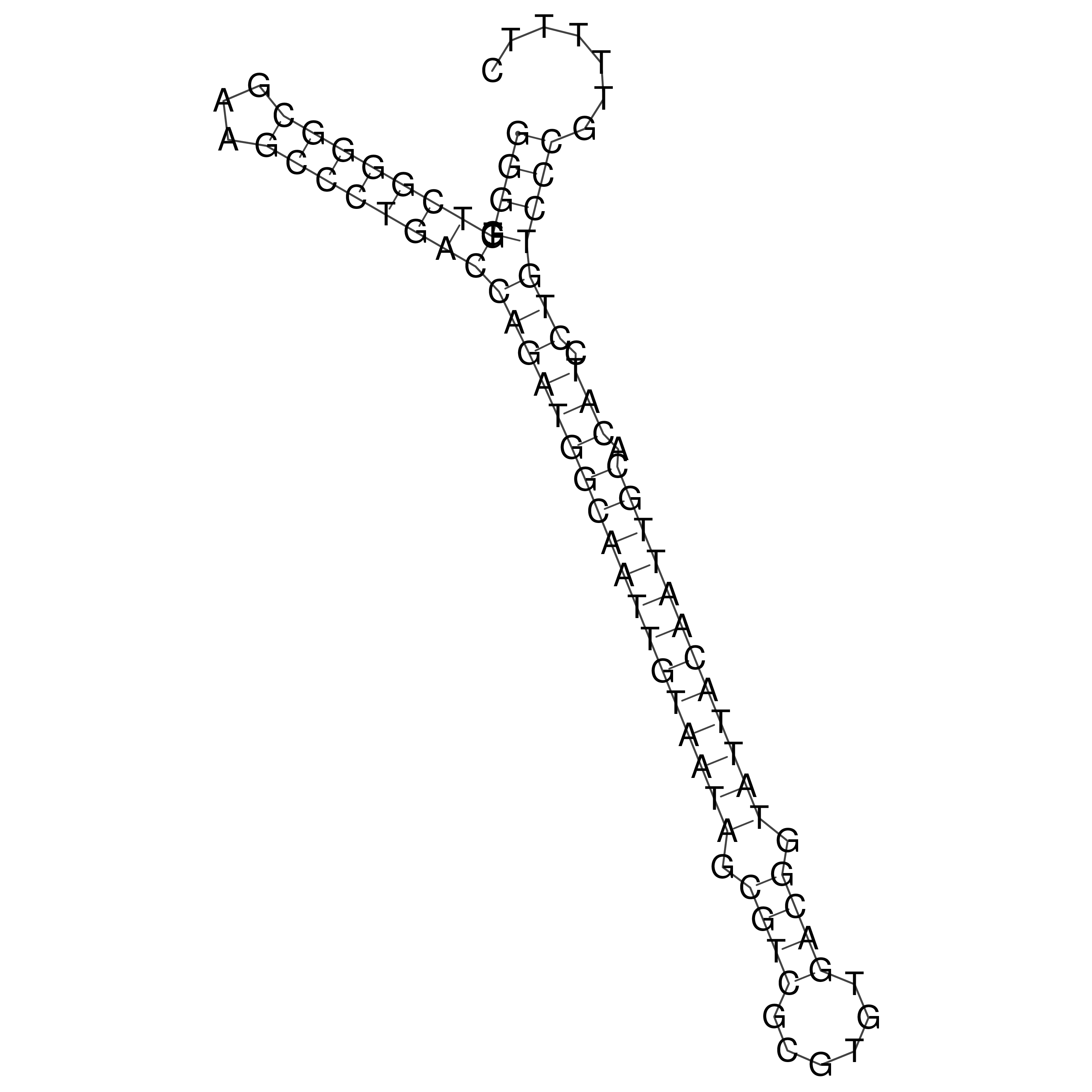
Relaxase
| ID | 9161 | GenBank | WP_107973980 |
| Name | nikB_E5O12_RS23595_p12-6919.1 |
UniProt ID | _ |
| Length | 899 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 104015.43 Da Isoelectric Point: 7.3623
>WP_107973980.1 IncI1-type relaxase NikB [Salmonella enterica]
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSHMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDKSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSHMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDKSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 5351 | GenBank | WP_052938717 |
| Name | WP_052938717_p12-6919.1 |
UniProt ID | A0A706B4V0 |
| Length | 110 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 110 a.a. Molecular weight: 12643.59 Da Isoelectric Point: 10.7463
>WP_052938717.1 MULTISPECIES: IncI1-type relaxosome accessory protein NikA [Enterobacteriaceae]
MSDSTVRKKSEVRQKTVVRTLRFSPVEDETIRKKAEDSGLTVSAYIRNAALNKRINSRTDDAFLKELMRL
GRMQKHLFVQGKRTGDKEYAEVLVAITELTNTLRKQLMEG
MSDSTVRKKSEVRQKTVVRTLRFSPVEDETIRKKAEDSGLTVSAYIRNAALNKRINSRTDDAFLKELMRL
GRMQKHLFVQGKRTGDKEYAEVLVAITELTNTLRKQLMEG
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A706B4V0 |
T4CP
| ID | 10609 | GenBank | WP_001289275 |
| Name | trbC_E5O12_RS23590_p12-6919.1 |
UniProt ID | _ |
| Length | 763 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 763 a.a. Molecular weight: 86957.15 Da Isoelectric Point: 6.7713
>WP_001289275.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Enterobacteriaceae]
MSEHRVNPELLHRTAWGNPVWNALQSLNIYGFCLVASLVASFIWPLALPACLLFTLITMLVFSLQRWRCP
LRMPMTLECADPSQDRMIKRSLFSFWPTLFQYEVILESPASGIFYVGYQRVRDIGRELWLSMDDLTRHIM
FFATTGGGKTETIFAWAINPLCWARGFTLVDGKAQNDTARTIWYLARRFGREDDVEVINFMNGGKSRSEI
ILSGEKTRPQSNTWNPFCYSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDLSLVRTPSAWTEEPRKQHAYLSGQFSE
TFSTFTEAFGDIFAEDSGDIDIRDSIHSDRILMVMIPALDTSAHTTSALGRMFITQKSMILARDLGYRLE
GTDSDALEVKKYKGRFPYLCFLDEVGAYYTDRIAVEATQVRSLDFALILMAQDQERIEGQTTATNTATLM
QNTGTKFAGRIVSEGSTARTLKSAAGEEARARMNNLQRQDGIFGESWIDSPQISILMESKINVQELIELH
PGEFFSIFRGETVPSASFFIPDDEKSCSSDPVVINRYISVDAPRLDRLRRLVPRTTQRRIPSPENVSAII
GVLTAKPSRKRRKIRTEPHTIVDTFQQRIAGRQAAMAMLEEYDTDINARESALWETAVNTLKTTTREERR
IRYITLNRPELPETKEENQISVRAERAGINLLTLPQDNNHLTGRPVNGFHHKKTNRPDWDGMY
MSEHRVNPELLHRTAWGNPVWNALQSLNIYGFCLVASLVASFIWPLALPACLLFTLITMLVFSLQRWRCP
LRMPMTLECADPSQDRMIKRSLFSFWPTLFQYEVILESPASGIFYVGYQRVRDIGRELWLSMDDLTRHIM
FFATTGGGKTETIFAWAINPLCWARGFTLVDGKAQNDTARTIWYLARRFGREDDVEVINFMNGGKSRSEI
ILSGEKTRPQSNTWNPFCYSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDLSLVRTPSAWTEEPRKQHAYLSGQFSE
TFSTFTEAFGDIFAEDSGDIDIRDSIHSDRILMVMIPALDTSAHTTSALGRMFITQKSMILARDLGYRLE
GTDSDALEVKKYKGRFPYLCFLDEVGAYYTDRIAVEATQVRSLDFALILMAQDQERIEGQTTATNTATLM
QNTGTKFAGRIVSEGSTARTLKSAAGEEARARMNNLQRQDGIFGESWIDSPQISILMESKINVQELIELH
PGEFFSIFRGETVPSASFFIPDDEKSCSSDPVVINRYISVDAPRLDRLRRLVPRTTQRRIPSPENVSAII
GVLTAKPSRKRRKIRTEPHTIVDTFQQRIAGRQAAMAMLEEYDTDINARESALWETAVNTLKTTTREERR
IRYITLNRPELPETKEENQISVRAERAGINLLTLPQDNNHLTGRPVNGFHHKKTNRPDWDGMY
Protein domains
No domain identified.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 55194..87070
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| E5O12_RS23970 (E5O12_23970) | 50256..51323 | + | 1068 | WP_074182768 | type IV pilus biogenesis lipoprotein PilL | - |
| E5O12_RS23975 (E5O12_23975) | 51323..51760 | + | 438 | WP_000539807 | type IV pilus biogenesis protein PilM | - |
| E5O12_RS23980 (E5O12_23980) | 51774..53456 | + | 1683 | WP_094307684 | PilN family type IVB pilus formation outer membrane protein | - |
| E5O12_RS23985 (E5O12_23985) | 53449..54744 | + | 1296 | WP_064148456 | type 4b pilus protein PilO2 | - |
| E5O12_RS23990 (E5O12_23990) | 54731..55183 | + | 453 | WP_001247337 | type IV pilus biogenesis protein PilP | - |
| E5O12_RS23995 (E5O12_23995) | 55194..56747 | + | 1554 | WP_136614768 | GspE/PulE family protein | virB11 |
| E5O12_RS24000 (E5O12_24000) | 56760..57845 | + | 1086 | WP_063103211 | type II secretion system F family protein | - |
| E5O12_RS24005 (E5O12_24005) | 57862..58476 | + | 615 | WP_064148457 | type 4 pilus major pilin | - |
| E5O12_RS24010 (E5O12_24010) | 58486..59046 | + | 561 | WP_063124165 | lytic transglycosylase domain-containing protein | virB1 |
| E5O12_RS24015 (E5O12_24015) | 59031..59687 | + | 657 | WP_074182772 | A24 family peptidase | - |
| E5O12_RS24020 (E5O12_24020) | 59687..60991 | + | 1305 | WP_064148473 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| E5O12_RS24035 (E5O12_24035) | 61632..62786 | + | 1155 | WP_001139955 | tyrosine-type recombinase/integrase | - |
| E5O12_RS24040 (E5O12_24040) | 62937..63761 | + | 825 | WP_001690630 | conjugal transfer protein TraE | traE |
| E5O12_RS24045 (E5O12_24045) | 63847..65049 | + | 1203 | WP_000976353 | conjugal transfer protein TraF | - |
| E5O12_RS24050 (E5O12_24050) | 65109..65693 | + | 585 | WP_000977525 | histidine phosphatase family protein | - |
| E5O12_RS24055 (E5O12_24055) | 66088..66546 | + | 459 | WP_001079808 | IncI1-type conjugal transfer lipoprotein TraH | - |
| E5O12_RS24060 (E5O12_24060) | 66543..67361 | + | 819 | WP_032220737 | IncI1-type conjugal transfer lipoprotein TraI | traI |
| E5O12_RS24065 (E5O12_24065) | 67358..68506 | + | 1149 | WP_001024972 | plasmid transfer ATPase TraJ | virB11 |
| E5O12_RS24070 (E5O12_24070) | 68503..68793 | + | 291 | WP_001299214 | hypothetical protein | traK |
| E5O12_RS24075 (E5O12_24075) | 68808..69359 | + | 552 | WP_000014586 | phospholipase D family protein | - |
| E5O12_RS24080 (E5O12_24080) | 69449..73213 | + | 3765 | WP_021497833 | LPD7 domain-containing protein | - |
| E5O12_RS24085 (E5O12_24085) | 73231..73578 | + | 348 | WP_001055900 | conjugal transfer protein | traL |
| E5O12_RS24090 (E5O12_24090) | 73575..74267 | + | 693 | WP_000138551 | DotI/IcmL family type IV secretion protein | traM |
| E5O12_RS24095 (E5O12_24095) | 74278..75261 | + | 984 | WP_052938673 | IncI1-type conjugal transfer protein TraN | traN |
| E5O12_RS24100 (E5O12_24100) | 75264..76553 | + | 1290 | WP_001271991 | conjugal transfer protein TraO | traO |
| E5O12_RS24105 (E5O12_24105) | 76553..77257 | + | 705 | WP_001493811 | IncI1-type conjugal transfer protein TraP | traP |
| E5O12_RS24110 (E5O12_24110) | 77257..77784 | + | 528 | WP_001055569 | conjugal transfer protein TraQ | traQ |
| E5O12_RS24115 (E5O12_24115) | 77835..78239 | + | 405 | WP_021497835 | IncI1-type conjugal transfer protein TraR | traR |
| E5O12_RS24120 (E5O12_24120) | 78303..78491 | + | 189 | WP_001277255 | putative conjugal transfer protein TraS | - |
| E5O12_RS24125 (E5O12_24125) | 78475..79275 | + | 801 | WP_054170916 | IncI1-type conjugal transfer protein TraT | traT |
| E5O12_RS24130 (E5O12_24130) | 79365..82409 | + | 3045 | WP_001024780 | IncI1-type conjugal transfer protein TraU | traU |
| E5O12_RS24135 (E5O12_24135) | 82409..83023 | + | 615 | WP_000337400 | IncI1-type conjugal transfer protein TraV | traV |
| E5O12_RS24140 (E5O12_24140) | 82990..84192 | + | 1203 | WP_001189156 | IncI1-type conjugal transfer protein TraW | traW |
| E5O12_RS24145 (E5O12_24145) | 84221..84805 | + | 585 | WP_001037985 | IncI1-type conjugal transfer protein TraX | - |
| E5O12_RS24150 (E5O12_24150) | 84902..87070 | + | 2169 | WP_000698368 | DotA/TraY family protein | traY |
| E5O12_RS24155 (E5O12_24155) | 87141..87803 | + | 663 | WP_000644794 | plasmid IncI1-type surface exclusion protein ExcA | - |
| E5O12_RS24160 (E5O12_24160) | 87901..88110 | + | 210 | WP_032178830 | hemolysin expression modulator Hha | - |
| E5O12_RS24165 (E5O12_24165) | 88442..88846 | - | 405 | WP_001175009 | IS200/IS605 family transposase | - |
| E5O12_RS24170 (E5O12_24170) | 88905..90131 | + | 1227 | WP_024186316 | RNA-guided endonuclease TnpB family protein | - |
| E5O12_RS24655 | 90176..90313 | + | 138 | WP_047088497 | hypothetical protein | - |
| E5O12_RS24850 | 90378..90473 | - | 96 | WP_001303310 | DinQ-like type I toxin DqlB | - |
Host bacterium
| ID | 14729 | GenBank | NZ_CP039604 |
| Plasmid name | p12-6919.1 | Incompatibility group | IncI |
| Plasmid size | 90972 bp | Coordinate of oriT [Strand] | 8443..8531 [+] |
| Host baterium | Salmonella enterica subsp. enterica serovar 14[5]12:i:- strain PNCS014868 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |