Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114278 |
| Name | oriT_pcfr-XZ03 |
| Organism | Staphylococcus capitis strain XZ03 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP077712 (7922..7957 [+], 36 nt) |
| oriT length | 36 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 36 nt
>oriT_pcfr-XZ03
CACGCGATCGGAACGTTCGCATAAGTGCACCCTTAC
CACGCGATCGGAACGTTCGCATAAGTGCACCCTTAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file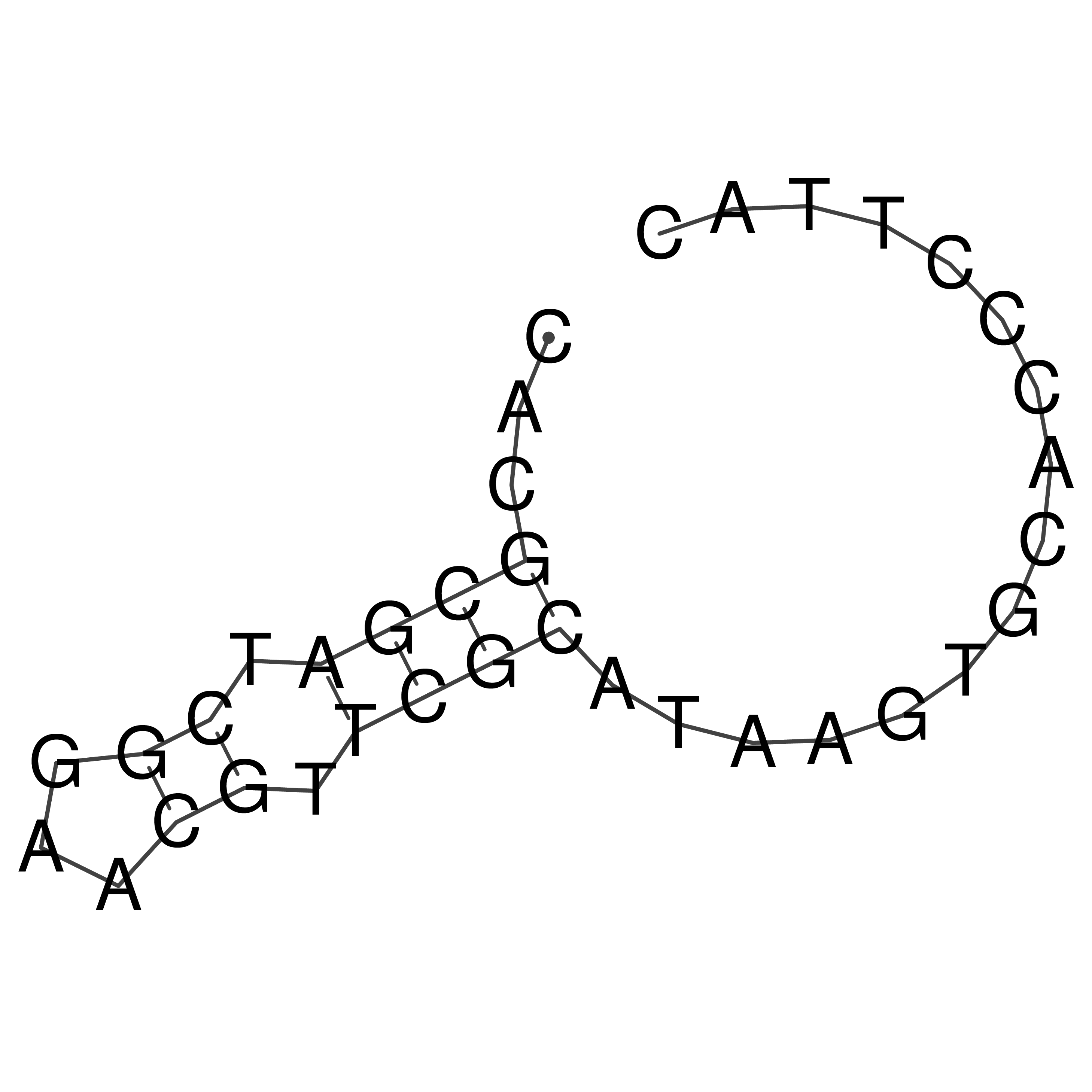
Relaxase
| ID | 9159 | GenBank | WP_030061449 |
| Name | MobA_MobL_KSP37_RS00035_pcfr-XZ03 |
UniProt ID | A0A848EWD6 |
| Length | 665 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 665 a.a. Molecular weight: 78725.60 Da Isoelectric Point: 5.1613
>WP_030061449.1 MULTISPECIES: MobA/MobL family protein [Staphylococcus]
MAMYHFQNKFVSKANGQSATAKAAYNSASRIKDFKENEFKDYSNKQCDYSEILLPNNADDKFKDREYLWN
KVNDVENRKNSQVAREIIIGLPNEFDPNSNIELAKEFAESLSNEGMIVDLNIHKINDENPHAHLLCTLRG
LDKNNEFEPKRKGNDYIRDWNTKDKHNEWRKRWETVQNKHLEMNGFSVRVSADSYKKQNIDLEPTKKEGW
KARKFEDETGKKSSISKHNESIKKRNQQKINKMFDEVDDMKSHKLNAFSYMDKSDSTTLKDMAKDLKIYV
TPINMYEENERLYDLKQKTALITDDEDRLNKIENIEDRQKKLESINEVFEKQAGIFFDKNYPEQALNYSD
DEKIFITRTILNDRDVLPANHELEDIVKEKRIKEAQISLNTVIGNRDISIESIEKESNFFADKLSNILDK
NNLSFDDVLENKHEGIEDSLKIDYYTSKLEVLRNAENTLEDYYDVQIKELFTDDEDYKAFNEVTDIKEKQ
QLIDFKTYHGTENTLEMLETGNFIPKYSDEDRKYITEQVKLLQEKEFKPNKNQHDKFVFGAIQKKLLNEY
DFDYSDNNDLKHLYQESNEVGDETSKDNIEEFYEGNDILVDKETYNNYNRKQQAYGLVNAGLDSMIFNFN
EIFRERMPKYINNQYKGKNHSKQRHELKNKRGMHL
MAMYHFQNKFVSKANGQSATAKAAYNSASRIKDFKENEFKDYSNKQCDYSEILLPNNADDKFKDREYLWN
KVNDVENRKNSQVAREIIIGLPNEFDPNSNIELAKEFAESLSNEGMIVDLNIHKINDENPHAHLLCTLRG
LDKNNEFEPKRKGNDYIRDWNTKDKHNEWRKRWETVQNKHLEMNGFSVRVSADSYKKQNIDLEPTKKEGW
KARKFEDETGKKSSISKHNESIKKRNQQKINKMFDEVDDMKSHKLNAFSYMDKSDSTTLKDMAKDLKIYV
TPINMYEENERLYDLKQKTALITDDEDRLNKIENIEDRQKKLESINEVFEKQAGIFFDKNYPEQALNYSD
DEKIFITRTILNDRDVLPANHELEDIVKEKRIKEAQISLNTVIGNRDISIESIEKESNFFADKLSNILDK
NNLSFDDVLENKHEGIEDSLKIDYYTSKLEVLRNAENTLEDYYDVQIKELFTDDEDYKAFNEVTDIKEKQ
QLIDFKTYHGTENTLEMLETGNFIPKYSDEDRKYITEQVKLLQEKEFKPNKNQHDKFVFGAIQKKLLNEY
DFDYSDNNDLKHLYQESNEVGDETSKDNIEEFYEGNDILVDKETYNNYNRKQQAYGLVNAGLDSMIFNFN
EIFRERMPKYINNQYKGKNHSKQRHELKNKRGMHL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A848EWD6 |
T4CP
| ID | 10607 | GenBank | WP_030061486 |
| Name | t4cp2_KSP37_RS00125_pcfr-XZ03 |
UniProt ID | _ |
| Length | 548 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 548 a.a. Molecular weight: 62899.75 Da Isoelectric Point: 5.9714
>WP_030061486.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Staphylococcaceae]
MTTLLGELESKATSINKKNLVIQDFDTKCPNRNIFVVGGPGSFKSAGYVIPNVIVKNQQSIVVTDPSGEV
YEKTANIKRMQGYDVRVVNFKNFLASDRQNFFDYIDKDSDCSIVAELIIKSAGDSKMNKDVWYKSSVGLL
NSLLLYAKYEFEPEKRTIGNIIKFIQNNKPNHDDEGSVELDNRFNELPKDHPARESYEFGFAVSEGRTRA
SIILTFVADLRNYIDNEIRSYTSDSDFDLRDVGLRKTIIYVMLPVLGNTVQSLSSLFFSQMFQQLYRLGD
ENGARLPVPVDFLLDEWPNIGAIPDYEETLATCRKYGISITTIVQSISQAVDKYNKDKANAIIGNHAVIL
CLGNVNNDTAKYIKEELGNATVEFETSSEGSSSGKDSRSKSSNKNVSYTGRALLNEDEISNMQQDKAILI
TKNRNPKIIKKLAYFKMFPNLTETYIAPQNEYKRKKNQSMIDKHNRLREEWDKKQEKIKQQKLEEYKEEQ
RQKELEEEQQAQEEQQNEEVKESKSKNEEQQEDKKDEQQKINDSKKAFYEKLKQKQAK
MTTLLGELESKATSINKKNLVIQDFDTKCPNRNIFVVGGPGSFKSAGYVIPNVIVKNQQSIVVTDPSGEV
YEKTANIKRMQGYDVRVVNFKNFLASDRQNFFDYIDKDSDCSIVAELIIKSAGDSKMNKDVWYKSSVGLL
NSLLLYAKYEFEPEKRTIGNIIKFIQNNKPNHDDEGSVELDNRFNELPKDHPARESYEFGFAVSEGRTRA
SIILTFVADLRNYIDNEIRSYTSDSDFDLRDVGLRKTIIYVMLPVLGNTVQSLSSLFFSQMFQQLYRLGD
ENGARLPVPVDFLLDEWPNIGAIPDYEETLATCRKYGISITTIVQSISQAVDKYNKDKANAIIGNHAVIL
CLGNVNNDTAKYIKEELGNATVEFETSSEGSSSGKDSRSKSSNKNVSYTGRALLNEDEISNMQQDKAILI
TKNRNPKIIKKLAYFKMFPNLTETYIAPQNEYKRKKNQSMIDKHNRLREEWDKKQEKIKQQKLEEYKEEQ
RQKELEEEQQAQEEQQNEEVKESKSKNEEQQEDKKDEQQKINDSKKAFYEKLKQKQAK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 13352..24955
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| KSP37_RS00045 (KSP37_00045) | 8422..8655 | + | 234 | WP_000602212 | hypothetical protein | - |
| KSP37_RS00050 (KSP37_00050) | 8936..9958 | + | 1023 | WP_224684886 | parM protein | - |
| KSP37_RS00055 (KSP37_00055) | 9961..10290 | + | 330 | WP_030061459 | plasmid segregation protein ParR | - |
| KSP37_RS00060 (KSP37_00060) | 10294..10554 | - | 261 | WP_001008214 | helix-turn-helix domain-containing protein | - |
| KSP37_RS00065 (KSP37_00065) | 10937..11896 | + | 960 | WP_030061463 | replication initiator protein A | - |
| KSP37_RS00070 (KSP37_00070) | 12000..12188 | - | 189 | WP_030061465 | hypothetical protein | - |
| KSP37_RS00075 (KSP37_00075) | 12361..13335 | + | 975 | WP_030061467 | conjugative transfer protein TrsA | - |
| KSP37_RS00080 (KSP37_00080) | 13352..13669 | + | 318 | WP_030061469 | CagC family type IV secretion system protein | virB2 |
| KSP37_RS00085 (KSP37_00085) | 13728..14135 | + | 408 | WP_030061471 | hypothetical protein | trsC |
| KSP37_RS00090 (KSP37_00090) | 14122..14805 | + | 684 | WP_030061473 | TrsD/TraD family conjugative transfer protein | trsD |
| KSP37_RS00095 (KSP37_00095) | 14826..16838 | + | 2013 | WP_030061475 | VirB4 family type IV secretion system protein | virb4 |
| KSP37_RS00100 (KSP37_00100) | 16850..18130 | + | 1281 | WP_030061477 | hypothetical protein | trsF |
| KSP37_RS00105 (KSP37_00105) | 18148..19224 | + | 1077 | WP_030061480 | CHAP domain-containing protein | trsG |
| KSP37_RS00110 (KSP37_00110) | 19234..19719 | + | 486 | WP_000735570 | TrsH/TraH family protein | - |
| KSP37_RS00115 (KSP37_00115) | 19735..21837 | + | 2103 | WP_030061483 | DNA topoisomerase III | - |
| KSP37_RS00120 (KSP37_00120) | 21853..22317 | + | 465 | WP_000715271 | hypothetical protein | trsJ |
| KSP37_RS00125 (KSP37_00125) | 22314..23960 | + | 1647 | WP_030061486 | VirD4-like conjugal transfer protein, CD1115 family | - |
| KSP37_RS00130 (KSP37_00130) | 24038..24955 | + | 918 | WP_030061488 | conjugal transfer protein TrbL family protein | trsL |
| KSP37_RS00135 (KSP37_00135) | 24972..25367 | + | 396 | WP_000608969 | single-stranded DNA-binding protein | - |
| KSP37_RS00140 (KSP37_00140) | 25424..26155 | + | 732 | WP_000869299 | hypothetical protein | - |
| KSP37_RS00145 (KSP37_00145) | 26192..26815 | + | 624 | WP_030061493 | hypothetical protein | - |
| KSP37_RS00150 (KSP37_00150) | 26831..27445 | + | 615 | WP_030061494 | hypothetical protein | - |
| KSP37_RS00155 (KSP37_00155) | 27514..27780 | + | 267 | WP_030061495 | hypothetical protein | - |
| KSP37_RS00160 (KSP37_00160) | 27866..29038 | + | 1173 | WP_032495458 | IS256-like element ISEnfa4 family transposase | - |
| KSP37_RS00165 (KSP37_00165) | 29041..29553 | + | 513 | WP_032495402 | MerR family transcriptional regulator | - |
| KSP37_RS00210 | 29766..29900 | + | 135 | WP_254903663 | hypothetical protein | - |
Host bacterium
| ID | 14713 | GenBank | NZ_CP077712 |
| Plasmid name | pcfr-XZ03 | Incompatibility group | - |
| Plasmid size | 39504 bp | Coordinate of oriT [Strand] | 7922..7957 [+] |
| Host baterium | Staphylococcus capitis strain XZ03 |
Cargo genes
| Drug resistance gene | cfr, aac(6')-aph(2'') |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |