Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114217 |
| Name | oriT_pTHAF9_e |
| Organism | Roseovarius sp. THAF9 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP045409 (1696..1715 [-], 20 nt) |
| oriT length | 20 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 20 nt
>oriT_pTHAF9_e
GTTTGCATAAGTGCGCCCTT
GTTTGCATAAGTGCGCCCTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file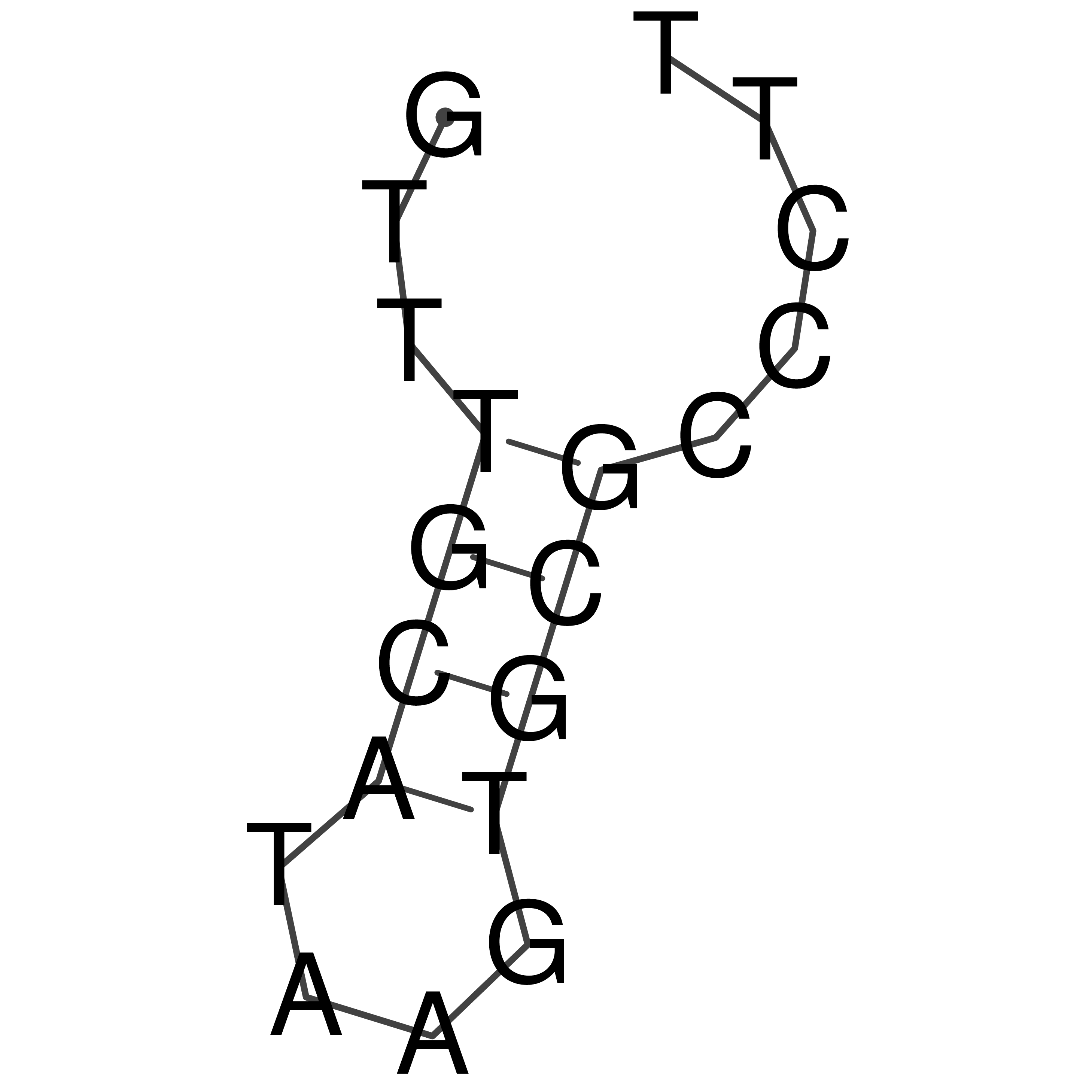
Relaxase
| ID | 9123 | GenBank | WP_037239043 |
| Name | mobQ_FIU86_RS22325_pTHAF9_e |
UniProt ID | A0A5P9I745 |
| Length | 430 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 430 a.a. Molecular weight: 48970.10 Da Isoelectric Point: 6.5830
>WP_037239043.1 MULTISPECIES: MobQ family relaxase [Roseovarius]
MASYHLSVKTIKRSAGRSATAAAAYRVGERIECLREGRVHDYTRKQGIEETFILTPKDAPDWAQDRSRLW
NEAEASETRRNSVTAREWELALPSEISDEDRSLITRQFAEELVSRYGVAVDVAIHAPHREGDQRNHHAHV
LTSTRKLEAGGFTAKTRILDSAKTGGVEIEQMRGLWAELQNRALERVGEVKRVDHRSLEKQRETALDRGD
KLSAEELDRDPELKLGPAANSMERRAKWMAERQGREYKPVTERGAVVHAARQARAAFREMRERLDIARET
YGMAREEGQGRVSAGLAALRAATAKDRSSERTPEDFQERLARVVDRSRDQDDTPKPDGRNYARELLKEIM
EKDAGRDGQAAVHKLDGHSDPELGEDTGREARKPSVNERLKDVLNKPRERLEPDNDRDHERDDDEQENTR
DRDRGEGHSL
MASYHLSVKTIKRSAGRSATAAAAYRVGERIECLREGRVHDYTRKQGIEETFILTPKDAPDWAQDRSRLW
NEAEASETRRNSVTAREWELALPSEISDEDRSLITRQFAEELVSRYGVAVDVAIHAPHREGDQRNHHAHV
LTSTRKLEAGGFTAKTRILDSAKTGGVEIEQMRGLWAELQNRALERVGEVKRVDHRSLEKQRETALDRGD
KLSAEELDRDPELKLGPAANSMERRAKWMAERQGREYKPVTERGAVVHAARQARAAFREMRERLDIARET
YGMAREEGQGRVSAGLAALRAATAKDRSSERTPEDFQERLARVVDRSRDQDDTPKPDGRNYARELLKEIM
EKDAGRDGQAAVHKLDGHSDPELGEDTGREARKPSVNERLKDVLNKPRERLEPDNDRDHERDDDEQENTR
DRDRGEGHSL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A5P9I745 |
T4CP
| ID | 10569 | GenBank | WP_037239039 |
| Name | t4cp2_FIU86_RS22315_pTHAF9_e |
UniProt ID | _ |
| Length | 557 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 557 a.a. Molecular weight: 61302.30 Da Isoelectric Point: 7.3026
>WP_037239039.1 type IV secretory system conjugative DNA transfer family protein [Roseovarius sp. THAF9]
MRGVMRVFGGLFRFFGRLIFAPILLGWMIGAVLFGAMIGALVATPFVFAFFDQPPGESAWQWLVFGPFIF
VGGVFGFQYWRMASGADAFFGLTGDSHGSARFANRKELKKLQREDGLLIGRNPHTGRLLHYGGPAHLITL
APTRAGKGVGTVIPNLLAAERSVLVIDPKGENARIAGEARRRFGTVHVLDPFEVSGMPSAAYNPLDRLAP
DSLDLGEDAASLTEALVMDPPGQVTEAHWNEEAKAILGGLIMFCVCHEDRNRRTLATVREYLTLPPEKLR
SLLELMQDSDAAGGLIARAANRFLGKADREAASVLSNAQRHTHFLDSPRIAKVLSRSDFHFSDLRHRITS
VFLVLPPNRMDAYSRWLRLLVSQALQDIARDAEASVRPLSGPADAQGGTQRLKTPTLFLLDEFAALGRLE
AVERAMGLMAGYGLQLWPILQDMSQLRDLYGERAGTFIANAGVQQVFGVNDFETAKWLSQMMGQETAGFQ
TDSFKPGDGPSFSNNLTGRDLLTPDEIMQLRPENQLLRVQGQATAVAQKLRYYADPEFKGLFVPQDQ
MRGVMRVFGGLFRFFGRLIFAPILLGWMIGAVLFGAMIGALVATPFVFAFFDQPPGESAWQWLVFGPFIF
VGGVFGFQYWRMASGADAFFGLTGDSHGSARFANRKELKKLQREDGLLIGRNPHTGRLLHYGGPAHLITL
APTRAGKGVGTVIPNLLAAERSVLVIDPKGENARIAGEARRRFGTVHVLDPFEVSGMPSAAYNPLDRLAP
DSLDLGEDAASLTEALVMDPPGQVTEAHWNEEAKAILGGLIMFCVCHEDRNRRTLATVREYLTLPPEKLR
SLLELMQDSDAAGGLIARAANRFLGKADREAASVLSNAQRHTHFLDSPRIAKVLSRSDFHFSDLRHRITS
VFLVLPPNRMDAYSRWLRLLVSQALQDIARDAEASVRPLSGPADAQGGTQRLKTPTLFLLDEFAALGRLE
AVERAMGLMAGYGLQLWPILQDMSQLRDLYGERAGTFIANAGVQQVFGVNDFETAKWLSQMMGQETAGFQ
TDSFKPGDGPSFSNNLTGRDLLTPDEIMQLRPENQLLRVQGQATAVAQKLRYYADPEFKGLFVPQDQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 14652 | GenBank | NZ_CP045409 |
| Plasmid name | pTHAF9_e | Incompatibility group | - |
| Plasmid size | 27503 bp | Coordinate of oriT [Strand] | 1696..1715 [-] |
| Host baterium | Roseovarius sp. THAF9 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |