Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114046 |
| Name | oriT_pYULZMPS10 |
| Organism | Salmonella enterica subsp. enterica serovar London strain LZ19MPS10 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP100354 (67797..67947 [+], 151 nt) |
| oriT length | 151 nt |
| IRs (inverted repeats) | 61..67, 73..79 (ATGCTCA..TGAGCAT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 151 nt
>oriT_pYULZMPS10
TGGTCGATGTGAACCCTTTTGACCGGGTTATGAATGAATTGAAAAATCGTGGCCGCAAAAATGCTCATATCCTGAGCATCCTTCAATTCGATTGGCCTGCATCAGAGGTCATCATCGAGAAGCTGAGCTGCTACATCACAGACGGTATTAA
TGGTCGATGTGAACCCTTTTGACCGGGTTATGAATGAATTGAAAAATCGTGGCCGCAAAAATGCTCATATCCTGAGCATCCTTCAATTCGATTGGCCTGCATCAGAGGTCATCATCGAGAAGCTGAGCTGCTACATCACAGACGGTATTAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file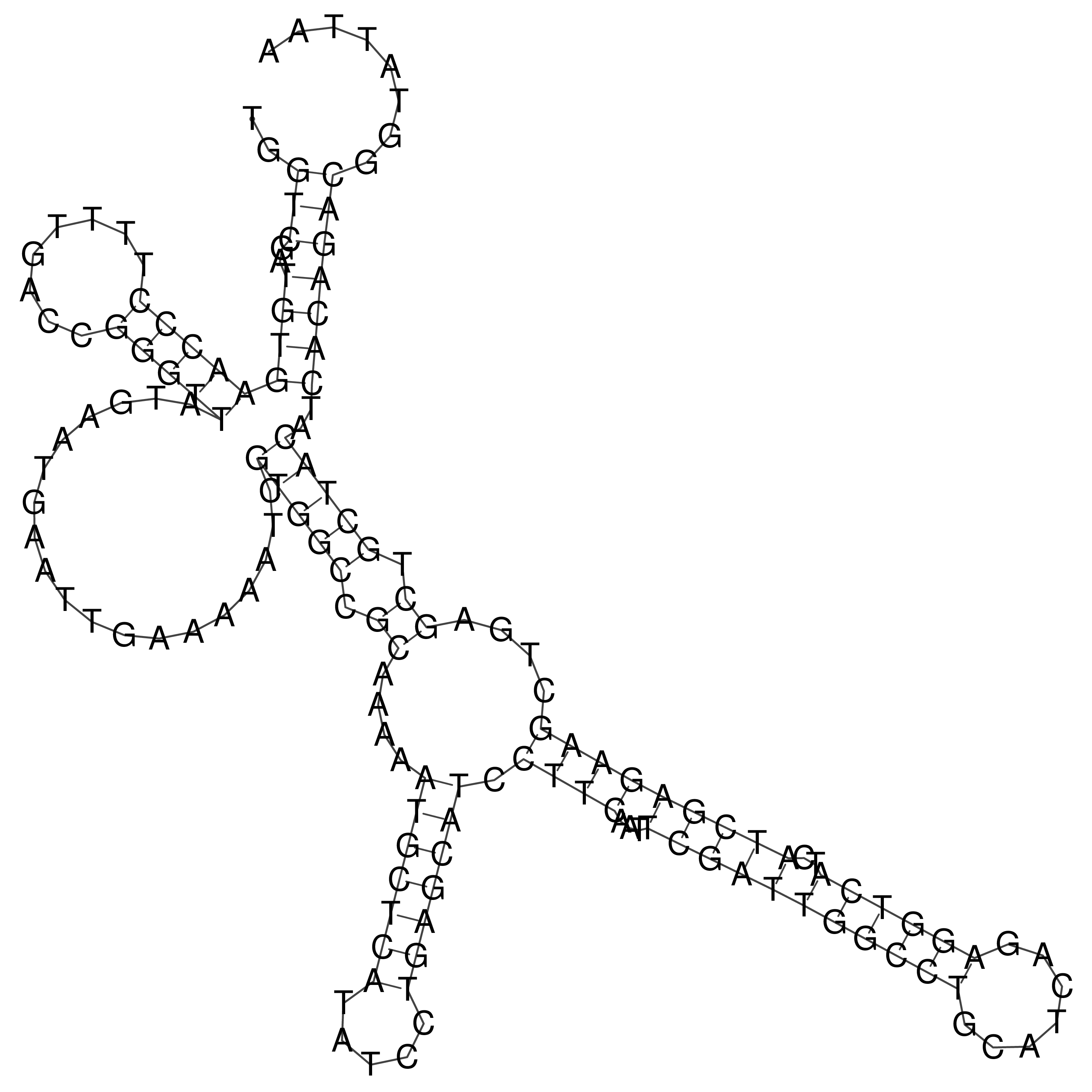
Relaxase
| ID | 9025 | GenBank | WP_176453579 |
| Name | mobH_NLL85_RS00430_pYULZMPS10 |
UniProt ID | _ |
| Length | 989 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 989 a.a. Molecular weight: 109277.46 Da Isoelectric Point: 4.7510
>WP_176453579.1 MULTISPECIES: MobH family relaxase [Enterobacteriaceae]
MLKALNKLFGGRSGVIEATPSARVLPLKDVEDEEIPRYPPFAKGLPVAPLDKILATQAELIEKVRNSLGF
TVDDFNRLVLPVIQRYAAFVHLLPASESHHHRGAGGLFRHGLEVAFWATQASESVIFSIEGTPRERRDNE
PRWRLASCFSGLLHDVGKPLSDVSITDKDGSITWNPYSESLHDWAHRHGIERYFIRWRDKRHKRHEQFSL
LAWHRIIPAETQEFLSKSGPSIIEAMLEAISGTTVNQPVTRLMLRADQESVSRDLRQSRLDVDEFSYGVP
VERYVFDAIRRLVKTGKWKVNEPGAKVWHLNQGVFIAWKQLGDLYDLISQDKIPGIPRDPDTLADILIER
GFAVPNTVQEKGERAYYRYWEVLPEMLQEATGSLKILMLRLESNDLVFTNEPPSAVAAEVVGDVEDAEIE
FVDPDESDDDQDEGGVSLNDDMLAAEQEAEKALAGLGFGDAMEMLKSTSTEPEDTPAQGAAETKDSPKAT
SATTPSKGKQVKAKPKKDTEEQTEKPEAKEELSPQDIAKNAPPLANDNPLQALKDVGGGLGDIDFPFDAF
NASTDACIADEESLKNSNEAELEQPKPDFVPEEQNSLQDDDFPMFSGSDEPPSWAIEPLPMLAVESEEAS
ATTEMSQANIPNQQEKDAKTLLSEMLAGYGEASALLEQAIMPVLEGKKTLGEVLCLMKGQAVILYPEGAR
SLGAPSEVLSKLSHANAIVPDPIMPGRKVRDFSGVKAIVLAQQLSDAVVAAIKEVEASMGGYQDVFELVS
PPDSDVRKSKSAPKQQSQPMQKKNQQQKSDAGSGQAAPAKDTKGKAAQKPQKEKKGDTSSVVAGHESKPI
EEKQNVARLPKREAQPEAVVVTKVEREKELGHIEVREREEQEVREFEPPKAKTNPKDIKDEDFLPPGVTP
EKAILILKDMIQKRSGRWLVTPVLEEDGCLVTSDKAFDMIAGENIGINKHILCGMLSRAQKRPLLKKRQG
KLYLEVDET
MLKALNKLFGGRSGVIEATPSARVLPLKDVEDEEIPRYPPFAKGLPVAPLDKILATQAELIEKVRNSLGF
TVDDFNRLVLPVIQRYAAFVHLLPASESHHHRGAGGLFRHGLEVAFWATQASESVIFSIEGTPRERRDNE
PRWRLASCFSGLLHDVGKPLSDVSITDKDGSITWNPYSESLHDWAHRHGIERYFIRWRDKRHKRHEQFSL
LAWHRIIPAETQEFLSKSGPSIIEAMLEAISGTTVNQPVTRLMLRADQESVSRDLRQSRLDVDEFSYGVP
VERYVFDAIRRLVKTGKWKVNEPGAKVWHLNQGVFIAWKQLGDLYDLISQDKIPGIPRDPDTLADILIER
GFAVPNTVQEKGERAYYRYWEVLPEMLQEATGSLKILMLRLESNDLVFTNEPPSAVAAEVVGDVEDAEIE
FVDPDESDDDQDEGGVSLNDDMLAAEQEAEKALAGLGFGDAMEMLKSTSTEPEDTPAQGAAETKDSPKAT
SATTPSKGKQVKAKPKKDTEEQTEKPEAKEELSPQDIAKNAPPLANDNPLQALKDVGGGLGDIDFPFDAF
NASTDACIADEESLKNSNEAELEQPKPDFVPEEQNSLQDDDFPMFSGSDEPPSWAIEPLPMLAVESEEAS
ATTEMSQANIPNQQEKDAKTLLSEMLAGYGEASALLEQAIMPVLEGKKTLGEVLCLMKGQAVILYPEGAR
SLGAPSEVLSKLSHANAIVPDPIMPGRKVRDFSGVKAIVLAQQLSDAVVAAIKEVEASMGGYQDVFELVS
PPDSDVRKSKSAPKQQSQPMQKKNQQQKSDAGSGQAAPAKDTKGKAAQKPQKEKKGDTSSVVAGHESKPI
EEKQNVARLPKREAQPEAVVVTKVEREKELGHIEVREREEQEVREFEPPKAKTNPKDIKDEDFLPPGVTP
EKAILILKDMIQKRSGRWLVTPVLEEDGCLVTSDKAFDMIAGENIGINKHILCGMLSRAQKRPLLKKRQG
KLYLEVDET
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 10455 | GenBank | WP_022635536 |
| Name | traD_NLL85_RS00435_pYULZMPS10 |
UniProt ID | _ |
| Length | 621 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 621 a.a. Molecular weight: 69430.92 Da Isoelectric Point: 7.1173
>WP_022635536.1 MULTISPECIES: conjugative transfer system coupling protein TraD [Gammaproteobacteria]
MTMSYDPLAYEMPWRPNYEKNAVAGWLAASGAALAVEQVSTMPPEPFYWMTGICGVMAMARLPKAIKLHL
LQKHLRGRDLEFISITELQKYIKDTPEDMWLGSGFLWENRHAQRVFEILKRDWTSIVGKESTVKKVVRKI
QGKRKELPIGQPWIHGVEPKEEKLMQPLKHTEGHTLIVGTTGSGKTRMFDILISQAILRGEAVIIIDPKG
DKEMRDNARRACEAMGQPERFVSFHPAFPEESVRIDPLRNFTRVTEIASRLAALIPSEAGADPFKSFGWQ
ALNNIAQGLVITHDRPNLTKLRRFLEGGAAGLVIRAVQAYSERVRPDWEAEAAPFLEKVKNGSREKIAFA
LMRFYYEIIQPEHPNSDLEGLLSMFQHDQTHFSKMVANLLPIMNMLTSGELGPLLSPDSTDLSDERQITD
SAKIINNAQVAYLGLDSLTDNMVGSAMGSIFLSDLTAVAGDRYNYGVNNRPVNIFVDEAAEVINDPFIQL
LNKGRGAKLRLFVATQTFADFAARLGSKDKALQVLGNINNTFALRIVDGETQEYIADNLPKTRLKYVMRT
QGQNSDGKEPIMHGGNQGERLMEEEADLFPAQLLGMLPNLEYIAKISGGTIVKGRLPILTQ
MTMSYDPLAYEMPWRPNYEKNAVAGWLAASGAALAVEQVSTMPPEPFYWMTGICGVMAMARLPKAIKLHL
LQKHLRGRDLEFISITELQKYIKDTPEDMWLGSGFLWENRHAQRVFEILKRDWTSIVGKESTVKKVVRKI
QGKRKELPIGQPWIHGVEPKEEKLMQPLKHTEGHTLIVGTTGSGKTRMFDILISQAILRGEAVIIIDPKG
DKEMRDNARRACEAMGQPERFVSFHPAFPEESVRIDPLRNFTRVTEIASRLAALIPSEAGADPFKSFGWQ
ALNNIAQGLVITHDRPNLTKLRRFLEGGAAGLVIRAVQAYSERVRPDWEAEAAPFLEKVKNGSREKIAFA
LMRFYYEIIQPEHPNSDLEGLLSMFQHDQTHFSKMVANLLPIMNMLTSGELGPLLSPDSTDLSDERQITD
SAKIINNAQVAYLGLDSLTDNMVGSAMGSIFLSDLTAVAGDRYNYGVNNRPVNIFVDEAAEVINDPFIQL
LNKGRGAKLRLFVATQTFADFAARLGSKDKALQVLGNINNTFALRIVDGETQEYIADNLPKTRLKYVMRT
QGQNSDGKEPIMHGGNQGERLMEEEADLFPAQLLGMLPNLEYIAKISGGTIVKGRLPILTQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 10456 | GenBank | WP_254155507 |
| Name | traC_NLL85_RS00520_pYULZMPS10 |
UniProt ID | _ |
| Length | 814 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 814 a.a. Molecular weight: 92236.42 Da Isoelectric Point: 5.9175
>WP_254155507.1 type IV secretion system protein TraC [Salmonella enterica]
MIVTIKKKLQETLIPEHLRAAGIIPVLAYDEDDHVFLMDDHSVGFGFMCEPLCGADEKVQERMNGFLNQE
FPSKATLQFVLFRSPDINQEMYRMMGLRDGFRHELLTSVIKERINFLQHHTTDRISAKTNKGTYDNGLIQ
DLKLFVTCKVPINNNNPSESELQNLAQLRTKVESSLQTVGLRPRTMTAVNYIRIMSTILNWGPDASWRHD
AVDWEMDKPICEQIFDYGTDVEVSKNGLRLGDYHAKVMSAKKLPDVFYFGDALTYAGDLSGGNSSIKENY
MVVTNVFFPEAESTKNTLERKRQFTVNQAYGPMLKFVPVLADKKESFDTLYESMKEGAKPVKISYSVVLF
APTKERVEAAAMAARNIWRESRFELMEDKFVALPMFLNCLPFCTDRDAVRDLFRYKTMTTEQAAVVLPVF
GEWKGTGTYHAALISRNGQLMSLSLHDSNTNKNLVIAAESGSGKSFLTNELIFSYLSEGAQVWVIDAGKS
YQKLSEMLNGDFVHFEEGTHVCLNPFELIQNYEDEEDAIVSLVCAMASAKGLLDEWQISALKQVLSRLWD
EKGKEMKVDDIAECCLEGEDMRLRDIGQQLYAFTSQGSYGKYFSRKNNVSFQNQFTVLELDELQGRKHLR
QVVLLQLIYQIQQEVFLGERNRKKVVIVDEAWDLLKEGEVSTFMEHAYRKFRKYGGSVVIATQSINDLYE
NAVGRAIAENSASMYLLGQTEETVESVKRSGRLTLSEGGFHTLKTVHTIQGVYSEIFIKSKSGMGVGRLI
VGDFQKLLYSTDPVDVNAIDQFVKQGMSIPEAIKAVMRNRKQAA
MIVTIKKKLQETLIPEHLRAAGIIPVLAYDEDDHVFLMDDHSVGFGFMCEPLCGADEKVQERMNGFLNQE
FPSKATLQFVLFRSPDINQEMYRMMGLRDGFRHELLTSVIKERINFLQHHTTDRISAKTNKGTYDNGLIQ
DLKLFVTCKVPINNNNPSESELQNLAQLRTKVESSLQTVGLRPRTMTAVNYIRIMSTILNWGPDASWRHD
AVDWEMDKPICEQIFDYGTDVEVSKNGLRLGDYHAKVMSAKKLPDVFYFGDALTYAGDLSGGNSSIKENY
MVVTNVFFPEAESTKNTLERKRQFTVNQAYGPMLKFVPVLADKKESFDTLYESMKEGAKPVKISYSVVLF
APTKERVEAAAMAARNIWRESRFELMEDKFVALPMFLNCLPFCTDRDAVRDLFRYKTMTTEQAAVVLPVF
GEWKGTGTYHAALISRNGQLMSLSLHDSNTNKNLVIAAESGSGKSFLTNELIFSYLSEGAQVWVIDAGKS
YQKLSEMLNGDFVHFEEGTHVCLNPFELIQNYEDEEDAIVSLVCAMASAKGLLDEWQISALKQVLSRLWD
EKGKEMKVDDIAECCLEGEDMRLRDIGQQLYAFTSQGSYGKYFSRKNNVSFQNQFTVLELDELQGRKHLR
QVVLLQLIYQIQQEVFLGERNRKKVVIVDEAWDLLKEGEVSTFMEHAYRKFRKYGGSVVIATQSINDLYE
NAVGRAIAENSASMYLLGQTEETVESVKRSGRLTLSEGGFHTLKTVHTIQGVYSEIFIKSKSGMGVGRLI
VGDFQKLLYSTDPVDVNAIDQFVKQGMSIPEAIKAVMRNRKQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 65881..90978
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| NLL85_RS00420 | 61915..62148 | - | 234 | WP_176453578 | hypothetical protein | - |
| NLL85_RS00425 | 62130..62747 | - | 618 | WP_071681756 | hypothetical protein | - |
| NLL85_RS00430 | 62915..65884 | + | 2970 | WP_176453579 | MobH family relaxase | - |
| NLL85_RS00435 | 65881..67746 | + | 1866 | WP_022635536 | conjugative transfer system coupling protein TraD | virb4 |
| NLL85_RS00440 | 67757..68341 | + | 585 | WP_031280871 | hypothetical protein | - |
| NLL85_RS00445 | 68298..68927 | + | 630 | WP_022635537 | DUF4400 domain-containing protein | tfc7 |
| NLL85_RS00450 | 68936..69382 | + | 447 | WP_071681752 | hypothetical protein | - |
| NLL85_RS00455 | 69392..69769 | + | 378 | WP_013141541 | hypothetical protein | - |
| NLL85_RS00460 | 69769..70431 | + | 663 | WP_254155563 | hypothetical protein | - |
| NLL85_RS00465 | 70612..70755 | + | 144 | WP_001275801 | hypothetical protein | - |
| NLL85_RS00470 | 70767..71129 | + | 363 | WP_231897578 | hypothetical protein | - |
| NLL85_RS00475 | 71141..71308 | + | 168 | WP_176453581 | hypothetical protein | - |
| NLL85_RS00480 | 71277..71558 | + | 282 | WP_000805625 | type IV conjugative transfer system protein TraL | traL |
| NLL85_RS00485 | 71555..72181 | + | 627 | WP_022635539 | TraE/TraK family type IV conjugative transfer system protein | traE |
| NLL85_RS00490 | 72165..73082 | + | 918 | WP_022635540 | type-F conjugative transfer system secretin TraK | traK |
| NLL85_RS00495 | 73082..74401 | + | 1320 | WP_071681777 | TraB/VirB10 family protein | traB |
| NLL85_RS00500 | 74398..74976 | + | 579 | WP_010845916 | type IV conjugative transfer system lipoprotein TraV | traV |
| NLL85_RS00505 | 74989..75372 | + | 384 | WP_089140452 | TraA family conjugative transfer protein | - |
| NLL85_RS00510 | 75576..81062 | + | 5487 | WP_254155564 | Ig-like domain-containing protein | - |
| NLL85_RS00515 | 81211..81918 | + | 708 | WP_089140450 | DsbC family protein | trbB |
| NLL85_RS00520 | 81915..84359 | + | 2445 | WP_254155507 | type IV secretion system protein TraC | virb4 |
| NLL85_RS00525 | 84374..84691 | + | 318 | WP_013141553 | hypothetical protein | - |
| NLL85_RS00530 | 84688..85218 | + | 531 | WP_022635546 | S26 family signal peptidase | - |
| NLL85_RS00535 | 85181..86446 | + | 1266 | WP_089140448 | TrbC family F-type conjugative pilus assembly protein | traW |
| NLL85_RS00540 | 86443..87114 | + | 672 | WP_071681744 | EAL domain-containing protein | - |
| NLL85_RS00545 | 87135..88118 | + | 984 | WP_240433690 | TraU family protein | traU |
| NLL85_RS00550 | 88234..90978 | + | 2745 | WP_119828715 | conjugal transfer mating pair stabilization protein TraN | traN |
| NLL85_RS00555 | 91016..91879 | - | 864 | WP_254155509 | hypothetical protein | - |
| NLL85_RS00560 | 92001..92642 | - | 642 | WP_041979271 | hypothetical protein | - |
| NLL85_RS00565 | 92936..93256 | + | 321 | WP_013141561 | hypothetical protein | - |
| NLL85_RS00570 | 93561..93746 | + | 186 | WP_231858731 | hypothetical protein | - |
| NLL85_RS00575 | 93966..94934 | + | 969 | WP_113620223 | AAA family ATPase | - |
| NLL85_RS00580 | 94945..95850 | + | 906 | WP_254155510 | hypothetical protein | - |
Host bacterium
| ID | 14481 | GenBank | NZ_CP100354 |
| Plasmid name | pYULZMPS10 | Incompatibility group | IncA/C2 |
| Plasmid size | 158615 bp | Coordinate of oriT [Strand] | 67797..67947 [+] |
| Host baterium | Salmonella enterica subsp. enterica serovar London strain LZ19MPS10 |
Cargo genes
| Drug resistance gene | mcr-4.3 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |