Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114043 |
| Name | oriT_2011N-4075|unnamed2 |
| Organism | Yersinia massiliensis strain 2011N-4075 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP054047 (9025..9112 [-], 88 nt) |
| oriT length | 88 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 88 nt
>oriT_2011N-4075|unnamed2
TACGGGACAGGATATGTTTTGTAGCACCGCCTGCACGCAGTCGGCCCTACAAAACGACCTGGTCAGGGCAGAGCCCCGACACCCGCTA
TACGGGACAGGATATGTTTTGTAGCACCGCCTGCACGCAGTCGGCCCTACAAAACGACCTGGTCAGGGCAGAGCCCCGACACCCGCTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file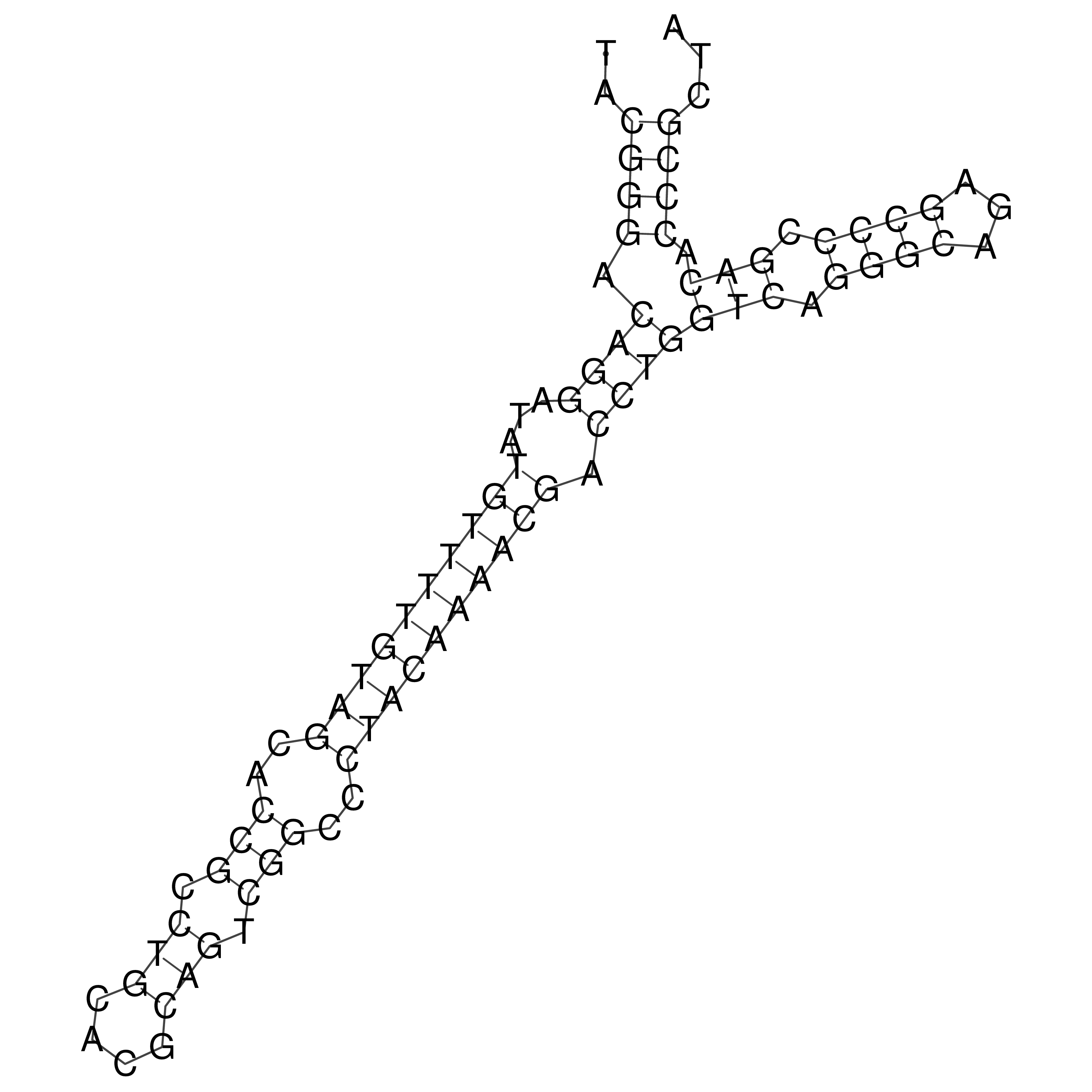
Relaxase
| ID | 9022 | GenBank | WP_172986752 |
| Name | traI_HRD68_RS00695_2011N-4075|unnamed2 |
UniProt ID | _ |
| Length | 651 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 651 a.a. Molecular weight: 73940.23 Da Isoelectric Point: 7.2343
>WP_172986752.1 TraI/MobA(P) family conjugative relaxase [Yersinia massiliensis]
MIPIVPEKRRDGRSSFIQLVSYLTLRDEVKPDMPISPDNPYIRPSRSKAAIFDRLIDYLDRKASVEEQTV
LAMFDDGTQQIKSGNVVCETNCFSLETASAEMNAVAMQNTRCVDAVYHCILSWREEDTPTDEQIFDSARY
CLKQLGMADHQYVVAVHRDTDNVHCHIAANRVNPKSYRATNLYNDVDTLHKACRHLEMKHGFTPDNGAWK
VNESHQVVRSQNDFKSIPRKAKQLEYYADSESLFSYATGECRQRIGEVVAGDNPSWDAIHTELIRAGLEL
KPKGEGLAIYSREDESLPPIKASSLHPDLTLYCLEETLGKFEPSPEIVTLTDDNDNIIHSNYKREFIYEP
RLHARDNTARIDRRLARADAREDLKARYQAYKSAWIKPKLEGGTLKQRYQNVAKRFAWQKARARIAIGDP
LLRKLTYHIIEVERMKAIAALRIAVKAEKKVFNANPENRRLSYRAWVEQQALNHDQAAIAQLRGWSYRLK
RANKTPPLSENGIRCAAADDCRPFNLEGYDTQINRDGVIQYRHDGIVQVQDRGERIEIANPFISDGQHIA
GAMTIAEEKSGEVMVFEGSRTFVNQACGLVGWFNEGGEKPLPLSDPQQRILAGYEVDRQASSSGGLQPQS
GTDQLHEREQGHKPQGGYRPQ
MIPIVPEKRRDGRSSFIQLVSYLTLRDEVKPDMPISPDNPYIRPSRSKAAIFDRLIDYLDRKASVEEQTV
LAMFDDGTQQIKSGNVVCETNCFSLETASAEMNAVAMQNTRCVDAVYHCILSWREEDTPTDEQIFDSARY
CLKQLGMADHQYVVAVHRDTDNVHCHIAANRVNPKSYRATNLYNDVDTLHKACRHLEMKHGFTPDNGAWK
VNESHQVVRSQNDFKSIPRKAKQLEYYADSESLFSYATGECRQRIGEVVAGDNPSWDAIHTELIRAGLEL
KPKGEGLAIYSREDESLPPIKASSLHPDLTLYCLEETLGKFEPSPEIVTLTDDNDNIIHSNYKREFIYEP
RLHARDNTARIDRRLARADAREDLKARYQAYKSAWIKPKLEGGTLKQRYQNVAKRFAWQKARARIAIGDP
LLRKLTYHIIEVERMKAIAALRIAVKAEKKVFNANPENRRLSYRAWVEQQALNHDQAAIAQLRGWSYRLK
RANKTPPLSENGIRCAAADDCRPFNLEGYDTQINRDGVIQYRHDGIVQVQDRGERIEIANPFISDGQHIA
GAMTIAEEKSGEVMVFEGSRTFVNQACGLVGWFNEGGEKPLPLSDPQQRILAGYEVDRQASSSGGLQPQS
GTDQLHEREQGHKPQGGYRPQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 10454 | GenBank | WP_254230646 |
| Name | trbC_HRD68_RS00680_2011N-4075|unnamed2 |
UniProt ID | _ |
| Length | 725 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 725 a.a. Molecular weight: 81712.34 Da Isoelectric Point: 6.4374
>WP_254230646.1 F-type conjugative transfer protein TrbC [Yersinia massiliensis]
MAQSVEIDARRVRRSLGYTFINDALLSPFGVQFFLLAGLVMGWIFPASLLISVPGLLLQVMLFSDRPFRM
PLRMPTDVGGIDLTTEREIPKYRNGLWGFFRYVVRTRKYLPAAGVMCLGFARGKALARELWLTLDDTLRH
MLLLATTGSGKTEALLSVFLNAICWGRGICYSDGKGQNSLALAMWSLARRFGREDDFYVLNFMTGGMDKL
LLLLLNDKKRQPSNTINLFGTANTTFIIQLMESMLPPAGSGDAGWQDKAKSMNAALVYAVYYKCKKEKRT
ISQTVIQSYLPLRKLAELYLEAKRDGWHKEAYNPLENYFNTLAGFRIELISQPSTWEQGVYDQHGYLIQQ
FNRMLTMFNDLYGHIFSTDAGDINIEDILHNDRILCTTIPALELSKGEASNIGKLYISAIRMTMARDLGC
ELEGMLNDVLIVKKYSGKFPFAIAMDELGAYFGPGMDNLASQMRSLGYMLIVSAQDIQRFIAEHRGEYMT
VNANLLTKWFMALQDEKDTFELAKITGGKGYYAELGAVEQAGGIITPNYEDATNNYIREKDRLDLGDLKE
LNPGEGMISFKSALVPSSAIYIPDDDKMTSALPMRINRFIEVQTPTEAELFALNPTLLRKLPPSAQEIED
ILQRIDEPEDSTHQIGIMDPILKRIVDLTLDLDNRADISYNPTQRGILLFEAAREALHQNKRKWRHQPTP
PKTIRVSKEVAISLADADLQNTISS
MAQSVEIDARRVRRSLGYTFINDALLSPFGVQFFLLAGLVMGWIFPASLLISVPGLLLQVMLFSDRPFRM
PLRMPTDVGGIDLTTEREIPKYRNGLWGFFRYVVRTRKYLPAAGVMCLGFARGKALARELWLTLDDTLRH
MLLLATTGSGKTEALLSVFLNAICWGRGICYSDGKGQNSLALAMWSLARRFGREDDFYVLNFMTGGMDKL
LLLLLNDKKRQPSNTINLFGTANTTFIIQLMESMLPPAGSGDAGWQDKAKSMNAALVYAVYYKCKKEKRT
ISQTVIQSYLPLRKLAELYLEAKRDGWHKEAYNPLENYFNTLAGFRIELISQPSTWEQGVYDQHGYLIQQ
FNRMLTMFNDLYGHIFSTDAGDINIEDILHNDRILCTTIPALELSKGEASNIGKLYISAIRMTMARDLGC
ELEGMLNDVLIVKKYSGKFPFAIAMDELGAYFGPGMDNLASQMRSLGYMLIVSAQDIQRFIAEHRGEYMT
VNANLLTKWFMALQDEKDTFELAKITGGKGYYAELGAVEQAGGIITPNYEDATNNYIREKDRLDLGDLKE
LNPGEGMISFKSALVPSSAIYIPDDDKMTSALPMRINRFIEVQTPTEAELFALNPTLLRKLPPSAQEIED
ILQRIDEPEDSTHQIGIMDPILKRIVDLTLDLDNRADISYNPTQRGILLFEAAREALHQNKRKWRHQPTP
PKTIRVSKEVAISLADADLQNTISS
Protein domains
No domain identified.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 53234..79109
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HRD68_RS00935 (HRD68_00935) | 49050..49472 | + | 423 | WP_172986791 | type IV pilus biogenesis protein PilM | - |
| HRD68_RS00940 (HRD68_00940) | 49498..51135 | + | 1638 | WP_172986792 | PilN family type IVB pilus formation outer membrane protein | - |
| HRD68_RS00945 (HRD68_00945) | 51356..52666 | + | 1311 | WP_172986793 | type 4b pilus protein PilO2 | - |
| HRD68_RS00950 (HRD68_00950) | 52653..53228 | + | 576 | WP_172986794 | type IV pilus biogenesis protein PilP | - |
| HRD68_RS00955 (HRD68_00955) | 53234..54724 | + | 1491 | WP_172986795 | GspE/PulE family protein | virB11 |
| HRD68_RS00960 (HRD68_00960) | 54721..55845 | + | 1125 | WP_172986796 | type II secretion system F family protein | - |
| HRD68_RS00965 (HRD68_00965) | 55881..56477 | + | 597 | WP_172986797 | type 4 pilus major pilin | - |
| HRD68_RS00970 (HRD68_00970) | 56532..57017 | + | 486 | WP_172986798 | lytic transglycosylase domain-containing protein | virB1 |
| HRD68_RS00975 (HRD68_00975) | 57017..57670 | + | 654 | WP_172986799 | A24 family peptidase | - |
| HRD68_RS00980 (HRD68_00980) | 57667..58857 | + | 1191 | WP_172986800 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| HRD68_RS00985 (HRD68_00985) | 59199..59867 | + | 669 | WP_172986801 | hypothetical protein | - |
| HRD68_RS00990 (HRD68_00990) | 59955..60410 | + | 456 | WP_172986802 | DotD/TraH family lipoprotein | - |
| HRD68_RS00995 (HRD68_00995) | 60407..61195 | + | 789 | WP_172986803 | type IV secretory system conjugative DNA transfer family protein | traI |
| HRD68_RS01000 (HRD68_01000) | 61215..62372 | + | 1158 | WP_172986804 | plasmid transfer ATPase TraJ | virB11 |
| HRD68_RS01005 (HRD68_01005) | 62372..62638 | + | 267 | WP_172986805 | IcmT/TraK family protein | traK |
| HRD68_RS01010 (HRD68_01010) | 62697..65885 | + | 3189 | WP_172986806 | LPD7 domain-containing protein | - |
| HRD68_RS01015 (HRD68_01015) | 65895..66380 | + | 486 | WP_172986807 | hypothetical protein | traL |
| HRD68_RS01020 (HRD68_01020) | 66465..67139 | + | 675 | WP_172986808 | DotI/IcmL/TraM family protein | traM |
| HRD68_RS01025 (HRD68_01025) | 67142..68152 | + | 1011 | WP_172986809 | DotH/IcmK family type IV secretion protein | traN |
| HRD68_RS01030 (HRD68_01030) | 68157..69383 | + | 1227 | WP_172986810 | conjugal transfer protein TraO | traO |
| HRD68_RS01035 (HRD68_01035) | 69383..70120 | + | 738 | WP_172986811 | hypothetical protein | traP |
| HRD68_RS01040 (HRD68_01040) | 70129..70659 | + | 531 | WP_172986812 | conjugal transfer protein TraQ | traQ |
| HRD68_RS01045 (HRD68_01045) | 70674..71078 | + | 405 | WP_172986813 | DUF6750 family protein | traR |
| HRD68_RS01050 (HRD68_01050) | 71170..71775 | + | 606 | WP_172986814 | hypothetical protein | traT |
| HRD68_RS01055 (HRD68_01055) | 71778..74852 | + | 3075 | WP_172986815 | ATP-binding protein | traU |
| HRD68_RS01060 (HRD68_01060) | 75043..76272 | + | 1230 | WP_172986816 | conjugal transfer protein TraW | traW |
| HRD68_RS01065 (HRD68_01065) | 76265..76864 | + | 600 | WP_172986817 | conjugal transfer protein TraX | - |
| HRD68_RS01070 (HRD68_01070) | 76998..79109 | + | 2112 | WP_254230645 | DotA/TraY family protein | traY |
Host bacterium
| ID | 14478 | GenBank | NZ_CP054047 |
| Plasmid name | 2011N-4075|unnamed2 | Incompatibility group | - |
| Plasmid size | 79154 bp | Coordinate of oriT [Strand] | 9025..9112 [-] |
| Host baterium | Yersinia massiliensis strain 2011N-4075 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |