Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114026 |
| Name | oriT_pDG3-1g |
| Organism | Agrobacterium salinitolerans strain DG3-1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP074400 (341001..341029 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pDG3-1g
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file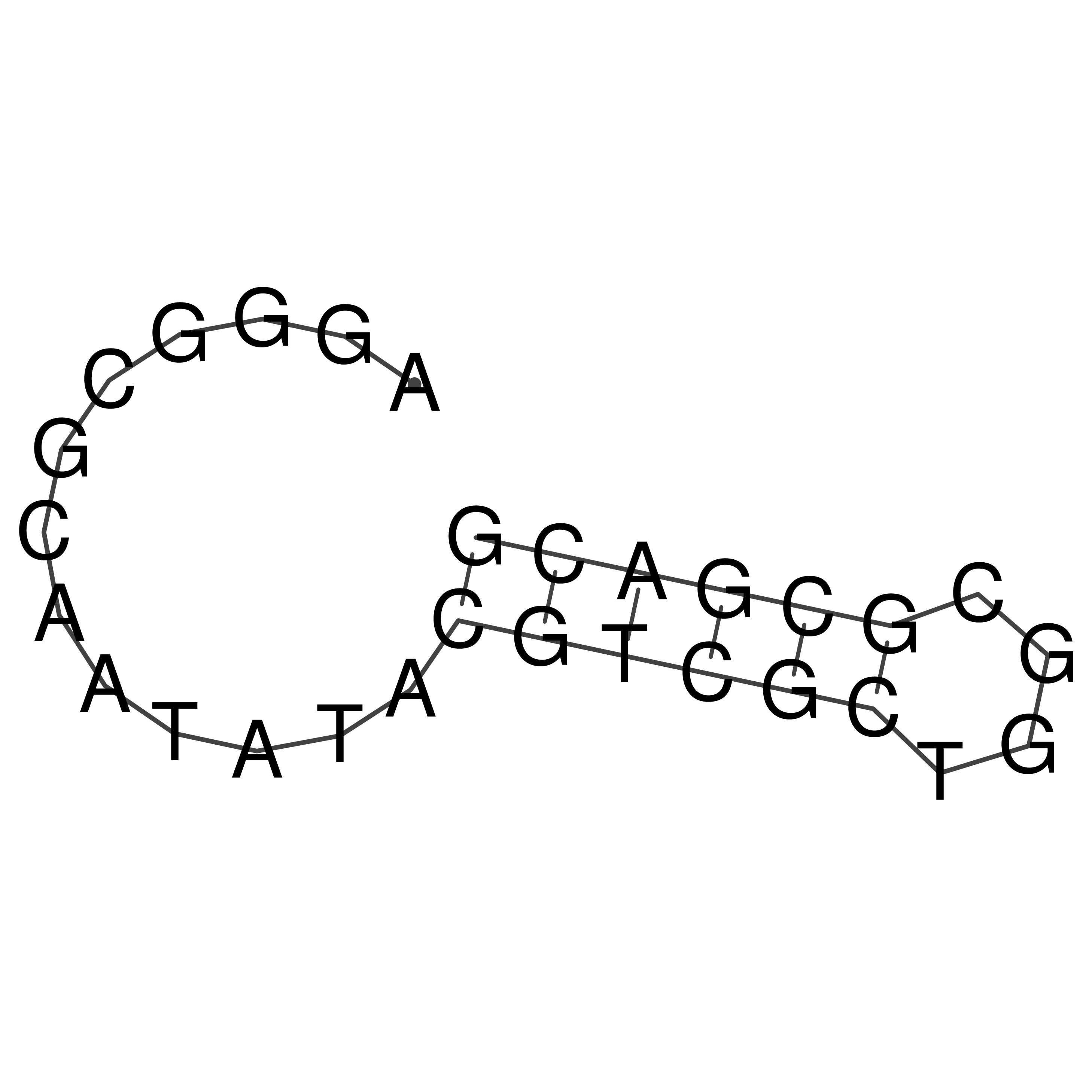
Relaxase
| ID | 9012 | GenBank | WP_217005982 |
| Name | traA_KHC17_RS28135_pDG3-1g |
UniProt ID | _ |
| Length | 1544 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1544 a.a. Molecular weight: 170896.51 Da Isoelectric Point: 9.4201
>WP_217005982.1 Ti-type conjugative transfer relaxase TraA [Agrobacterium salinitolerans]
MAIMFIRAQVISRGGGRSIVSAAAYRHRARMVDEQAGASFSYRGGAAELKHEELALPDQIPAWLRTAIDG
KSVAAASEALWNAVDAFETRADAQLARELIVALPEELTRSENIALVREFVRDNLTSRGMIADWVYHDKDG
NPHIHLMTALRPLADDGFGPKKVPVIGTNGEPMRVVTPDRPKGKIVYKLWAGDKETMKSWKIAWAETTNR
HLALAGHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIEMYFAPADLARRHEMADRLLAEPALLLKQ
LANERSTFDERDIARVLHRYVDDPTDFANIRAKLMASNDLVLLKPQQADLQTGEMAEAAVFTTRDILRAE
YDMAQSAEALALRKGFGVAAAKIAAAVRTIETGDAENRFRLDREQVEAVHHIAGDNGIAAVVGLAGAGKS
TLLAAARVAWESDGHRVFGAALAGKAAEGLEDSSGTKSRTLASWEMAWENGRDLLHRQDIFVIDEAGMVS
SHQMACVLRRVEEAGAKVVLVGDAMQLQPIQAGAAFRAISERIGSAELAGVRRQREEWARQASRLFARGQ
VENGLDAYAQRGHIVETGSRDEAVERIVTDWTSARRDLIQQNADKEQPARLRGGELLVLAHTNDDVRRLN
EALRNVMKDEAALTASREFQTARGVREFAGGDRIIFLENARFLEPRARKLGPQYVKNGMLGTVVSTGGLH
SGPLLSVRLDNGRDVVISENSYRNVDHGYAATIHKSQGASVDRTFVLATGMMDQHLAYVSMTRHRDRADL
YAAREDFKPKWEWGMRSPIDHAAGVTGELVGTGNAMFRPQDEGADKSPYADVKTDDGTVHRLWGVSLPKA
LDAAGVSEGDTVTLRKDGVERVTVTVPVIGEKTGKKRLEQRTVDRNVWTAKQIETAEVRQQRIEQESHRP
ELFKQLVERLGRSGAKTTTLDFESEAGYQAHARDFAHRRGIDTLSGVAAWMEAGAGRPLAWLAQKREQVA
KLWERASVALGFAIERERRFSYNEECTENLATQVPADRRYLMAPTTLFSRGVDEDARRAQLRSDRWKERD
AILQPVLQKIYRDPESALARLNALASDARIAPRELAEDLEASPQRLGRLRGSDILVDGRSARSERDTAVL
ALSELLPLARAHATEFRRQAERFGLREEQRRAHMSLSIPALSNQAMGRLAEIEAVRERGGTDAYKTAFTY
AAQDRLLVKEVRAVNDALTARFGWSAFTDKPDAVAERNMIERMPEDLHPDRREKLARLFAVVRRFAEEQH
LAETKDRSRIVAGASVEPGKETVGVLPMLAAVTEFETAIDQEARKRVRAVSHYGQNRTALFGAAERIWRD
PAGAVGKIEELILKGFAGERIAAAVANDPAAYGALRGSGRIMDKLLAVGRERKEALQAISEAASCIRSLG
VSWASAFDAETRAVSEERRCMSVAIPGLSQAAENALSTLVADINRSTGRQKKAGQDAVVRLLDPHIAREF
AEVSRALDERFGRNAILRGERDVINRVSPAQRRAFEAIKERLLVLQRAVREESSQKIIAERQRRSIERGR
IFTR
MAIMFIRAQVISRGGGRSIVSAAAYRHRARMVDEQAGASFSYRGGAAELKHEELALPDQIPAWLRTAIDG
KSVAAASEALWNAVDAFETRADAQLARELIVALPEELTRSENIALVREFVRDNLTSRGMIADWVYHDKDG
NPHIHLMTALRPLADDGFGPKKVPVIGTNGEPMRVVTPDRPKGKIVYKLWAGDKETMKSWKIAWAETTNR
HLALAGHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIEMYFAPADLARRHEMADRLLAEPALLLKQ
LANERSTFDERDIARVLHRYVDDPTDFANIRAKLMASNDLVLLKPQQADLQTGEMAEAAVFTTRDILRAE
YDMAQSAEALALRKGFGVAAAKIAAAVRTIETGDAENRFRLDREQVEAVHHIAGDNGIAAVVGLAGAGKS
TLLAAARVAWESDGHRVFGAALAGKAAEGLEDSSGTKSRTLASWEMAWENGRDLLHRQDIFVIDEAGMVS
SHQMACVLRRVEEAGAKVVLVGDAMQLQPIQAGAAFRAISERIGSAELAGVRRQREEWARQASRLFARGQ
VENGLDAYAQRGHIVETGSRDEAVERIVTDWTSARRDLIQQNADKEQPARLRGGELLVLAHTNDDVRRLN
EALRNVMKDEAALTASREFQTARGVREFAGGDRIIFLENARFLEPRARKLGPQYVKNGMLGTVVSTGGLH
SGPLLSVRLDNGRDVVISENSYRNVDHGYAATIHKSQGASVDRTFVLATGMMDQHLAYVSMTRHRDRADL
YAAREDFKPKWEWGMRSPIDHAAGVTGELVGTGNAMFRPQDEGADKSPYADVKTDDGTVHRLWGVSLPKA
LDAAGVSEGDTVTLRKDGVERVTVTVPVIGEKTGKKRLEQRTVDRNVWTAKQIETAEVRQQRIEQESHRP
ELFKQLVERLGRSGAKTTTLDFESEAGYQAHARDFAHRRGIDTLSGVAAWMEAGAGRPLAWLAQKREQVA
KLWERASVALGFAIERERRFSYNEECTENLATQVPADRRYLMAPTTLFSRGVDEDARRAQLRSDRWKERD
AILQPVLQKIYRDPESALARLNALASDARIAPRELAEDLEASPQRLGRLRGSDILVDGRSARSERDTAVL
ALSELLPLARAHATEFRRQAERFGLREEQRRAHMSLSIPALSNQAMGRLAEIEAVRERGGTDAYKTAFTY
AAQDRLLVKEVRAVNDALTARFGWSAFTDKPDAVAERNMIERMPEDLHPDRREKLARLFAVVRRFAEEQH
LAETKDRSRIVAGASVEPGKETVGVLPMLAAVTEFETAIDQEARKRVRAVSHYGQNRTALFGAAERIWRD
PAGAVGKIEELILKGFAGERIAAAVANDPAAYGALRGSGRIMDKLLAVGRERKEALQAISEAASCIRSLG
VSWASAFDAETRAVSEERRCMSVAIPGLSQAAENALSTLVADINRSTGRQKKAGQDAVVRLLDPHIAREF
AEVSRALDERFGRNAILRGERDVINRVSPAQRRAFEAIKERLLVLQRAVREESSQKIIAERQRRSIERGR
IFTR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 10446 | GenBank | WP_217005540 |
| Name | traG_KHC17_RS26585_pDG3-1g |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70900.99 Da Isoelectric Point: 9.3372
>WP_217005540.1 Ti-type conjugative transfer system protein TraG [Agrobacterium salinitolerans]
MKGKARLHPSLLLIIVPIVVTGFSFYVSHWRWPELANGITGKTQYWFLRASPLPVLLLGPLTGLLAVWVL
PLHRRRPVALASFLCFLLMAGFYGLRESGRLLPSLESGVLTWDRALSFVDMIAVVGTAMGFMVVAVAARI
SSIVPERVPRARRGTFGDADWLPMSAAARLFPDDGEIVIGERYRVDKELVHELPFDPNDPATWGRGGRAP
LLTYAQDFDSTHMLFFAGSGGFKTTSNVVPTALRYRGPLICLDPSTEVAPMVVDHRRDKLDREVVVLDPA
NPVMGFNVLDGIEHSLKKEEDIVGIAHMLLSESVRFESSTGSFFQNQAHNLLTGLLAHVMLSPEFAGRRN
LRSLRQIVSEPETSVLAMLRDVQEHSKSAFIRETLGVFVNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GNTFNSSEIAGGKKDVFINIPASILRSYPGIGRVIIGSLINAMIEADGAFTRRALFVLDEVDLLGYMRVL
EEARDRGRKYGVSMMLMYQSVGQLERHFGKDGAVSWIDGCAFVSYAAIKALDTARNVSAQCGEMTVEVKG
SSRNLGWGTKNSGSRKSESINFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMDMQAR
PNRFAKLGP
MKGKARLHPSLLLIIVPIVVTGFSFYVSHWRWPELANGITGKTQYWFLRASPLPVLLLGPLTGLLAVWVL
PLHRRRPVALASFLCFLLMAGFYGLRESGRLLPSLESGVLTWDRALSFVDMIAVVGTAMGFMVVAVAARI
SSIVPERVPRARRGTFGDADWLPMSAAARLFPDDGEIVIGERYRVDKELVHELPFDPNDPATWGRGGRAP
LLTYAQDFDSTHMLFFAGSGGFKTTSNVVPTALRYRGPLICLDPSTEVAPMVVDHRRDKLDREVVVLDPA
NPVMGFNVLDGIEHSLKKEEDIVGIAHMLLSESVRFESSTGSFFQNQAHNLLTGLLAHVMLSPEFAGRRN
LRSLRQIVSEPETSVLAMLRDVQEHSKSAFIRETLGVFVNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GNTFNSSEIAGGKKDVFINIPASILRSYPGIGRVIIGSLINAMIEADGAFTRRALFVLDEVDLLGYMRVL
EEARDRGRKYGVSMMLMYQSVGQLERHFGKDGAVSWIDGCAFVSYAAIKALDTARNVSAQCGEMTVEVKG
SSRNLGWGTKNSGSRKSESINFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMDMQAR
PNRFAKLGP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 10447 | GenBank | WP_217005978 |
| Name | traG_KHC17_RS28120_pDG3-1g |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70920.99 Da Isoelectric Point: 9.0337
>WP_217005978.1 Ti-type conjugative transfer system protein TraG [Agrobacterium salinitolerans]
MTGMARLHPSLLLIIVPIVVTGFSFYVSHWRWPELANGITGKTQYWFLRASPLPVLLLGPLAGLLAVWVL
PLHRRRPVALASFLCFLLMAGFYGLREFGRLLPSVESGVLTWDRALSFVDMIAVVGTAMGFMVVAVAARI
SSIVPERVPRARRGTFGDADWLPMSAAARLFPDDGEIVIGERYRVDKELVHELPFDPNDPATWGRGGRAP
LLTYAQDFDSTHMLFFAGSGGFKTTSNVVPTALRYRGPLICLDPSTEVAPMVVDHRRDKLDREVVVLDPA
NPVMGFNVLDGIEHSLKKEEDIVGIAHMLLSESVRFESSTGSFFQNQAHNLLTGLLAHVMLSPEFAGRRN
LRSLRQIVSEPETSVLAMLRDVQEHSRSAFIRETLGVFVNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GNTFNSSEIAGGKKDVFINIPASILRSYPGIGRVIIGSLINAMIEADGAFTRRALFVLDEVDLLGYMRVL
EEARDRGRKYGVSMMLMYQSVGQLERHFGKDGAVSWIDGCAFVSYAAIKALDTARNVSAQCGEMTVEVKG
SSRNLGWGTKNSGSRKSESINFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMDMQAR
PNRFAKLGP
MTGMARLHPSLLLIIVPIVVTGFSFYVSHWRWPELANGITGKTQYWFLRASPLPVLLLGPLAGLLAVWVL
PLHRRRPVALASFLCFLLMAGFYGLREFGRLLPSVESGVLTWDRALSFVDMIAVVGTAMGFMVVAVAARI
SSIVPERVPRARRGTFGDADWLPMSAAARLFPDDGEIVIGERYRVDKELVHELPFDPNDPATWGRGGRAP
LLTYAQDFDSTHMLFFAGSGGFKTTSNVVPTALRYRGPLICLDPSTEVAPMVVDHRRDKLDREVVVLDPA
NPVMGFNVLDGIEHSLKKEEDIVGIAHMLLSESVRFESSTGSFFQNQAHNLLTGLLAHVMLSPEFAGRRN
LRSLRQIVSEPETSVLAMLRDVQEHSRSAFIRETLGVFVNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GNTFNSSEIAGGKKDVFINIPASILRSYPGIGRVIIGSLINAMIEADGAFTRRALFVLDEVDLLGYMRVL
EEARDRGRKYGVSMMLMYQSVGQLERHFGKDGAVSWIDGCAFVSYAAIKALDTARNVSAQCGEMTVEVKG
SSRNLGWGTKNSGSRKSESINFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMDMQAR
PNRFAKLGP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 390214..399130
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| KHC17_RS28335 (KHC17_28335) | 386202..386750 | + | 549 | WP_217006041 | carboxymuconolactone decarboxylase family protein | - |
| KHC17_RS28340 (KHC17_28340) | 386819..387046 | + | 228 | Protein_389 | DUF1348 family protein | - |
| KHC17_RS28345 (KHC17_28345) | 387071..387418 | + | 348 | WP_217005578 | hypothetical protein | - |
| KHC17_RS28350 (KHC17_28350) | 387452..388342 | + | 891 | WP_217006043 | LysR family transcriptional regulator | - |
| KHC17_RS28355 (KHC17_28355) | 388867..389229 | - | 363 | WP_149148713 | transcriptional regulator | - |
| KHC17_RS29880 | 389323..389862 | + | 540 | WP_254229242 | hypothetical protein | - |
| KHC17_RS29885 | 389865..390217 | + | 353 | Protein_394 | hypothetical protein | - |
| KHC17_RS28365 (KHC17_28365) | 390214..390513 | + | 300 | WP_217006047 | TrbC/VirB2 family protein | virB2 |
| KHC17_RS28370 (KHC17_28370) | 390516..390857 | + | 342 | WP_149148710 | type IV secretion system protein VirB3 | virB3 |
| KHC17_RS28375 (KHC17_28375) | 390850..393231 | + | 2382 | WP_217006049 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| KHC17_RS28380 (KHC17_28380) | 393231..393929 | + | 699 | WP_217006051 | P-type DNA transfer protein VirB5 | virB5 |
| KHC17_RS28385 (KHC17_28385) | 393926..394159 | + | 234 | WP_217006054 | EexN family lipoprotein | - |
| KHC17_RS28390 (KHC17_28390) | 394164..395099 | + | 936 | WP_217006056 | type IV secretion system protein | virB6 |
| KHC17_RS28395 (KHC17_28395) | 395135..395401 | + | 267 | WP_217006058 | hypothetical protein | - |
| KHC17_RS28400 (KHC17_28400) | 395405..396076 | + | 672 | WP_217006060 | virB8 family protein | virB8 |
| KHC17_RS28405 (KHC17_28405) | 396073..396927 | + | 855 | WP_217006062 | P-type conjugative transfer protein VirB9 | virB9 |
| KHC17_RS28410 (KHC17_28410) | 396942..398114 | + | 1173 | WP_217006064 | type IV secretion system protein VirB10 | virB10 |
| KHC17_RS28415 (KHC17_28415) | 398123..399130 | + | 1008 | WP_217006066 | P-type DNA transfer ATPase VirB11 | virB11 |
| KHC17_RS28420 (KHC17_28420) | 399055..399372 | - | 318 | WP_333781944 | leucine zipper domain-containing protein | - |
| KHC17_RS28425 (KHC17_28425) | 399441..400586 | - | 1146 | WP_217006070 | HlyD family secretion protein | - |
| KHC17_RS28430 (KHC17_28430) | 400723..401622 | + | 900 | WP_217006072 | LysR family transcriptional regulator | - |
| KHC17_RS28435 (KHC17_28435) | 401702..403327 | - | 1626 | WP_217006074 | MFS transporter | - |
| KHC17_RS28440 (KHC17_28440) | 403602..404048 | - | 447 | WP_217006076 | carboxymuconolactone decarboxylase family protein | - |
Host bacterium
| ID | 14461 | GenBank | NZ_CP074400 |
| Plasmid name | pDG3-1g | Incompatibility group | - |
| Plasmid size | 521982 bp | Coordinate of oriT [Strand] | 341001..341029 [+] |
| Host baterium | Agrobacterium salinitolerans strain DG3-1 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |