Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114025 |
| Name | oriT_pDG3-1c |
| Organism | Agrobacterium salinitolerans strain DG3-1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP074396 (37246..37290 [+], 45 nt) |
| oriT length | 45 nt |
| IRs (inverted repeats) | 22..27, 32..37 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 45 nt
>oriT_pDG3-1c
GGTTCCAAGGGCGCAATTATACGTCGCTCACGCGACGCCTTGCGT
GGTTCCAAGGGCGCAATTATACGTCGCTCACGCGACGCCTTGCGT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file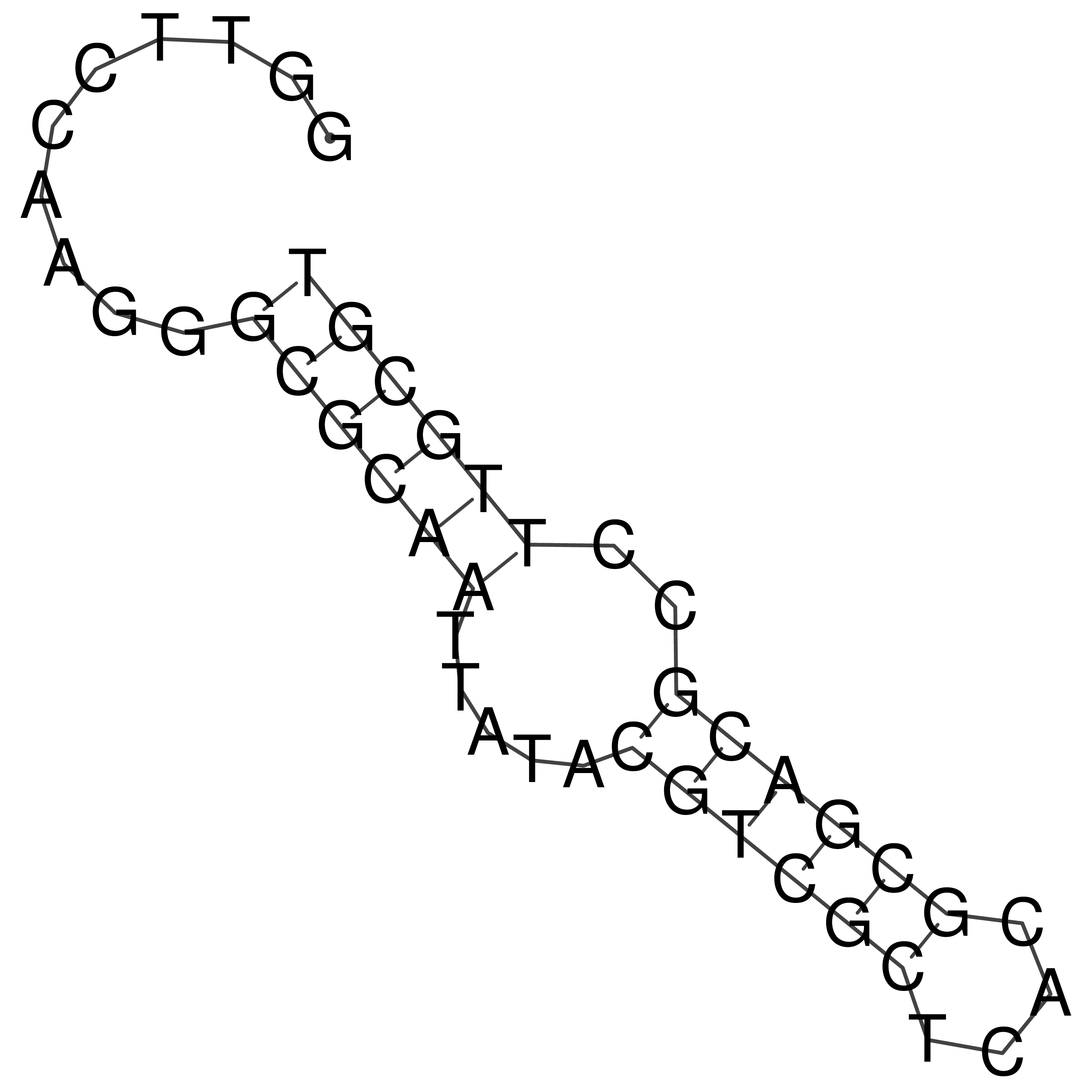
Relaxase
| ID | 9011 | GenBank | WP_217005269 |
| Name | traA_KHC17_RS24880_pDG3-1c |
UniProt ID | _ |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 123627.05 Da Isoelectric Point: 9.8722
>WP_217005269.1 Ti-type conjugative transfer relaxase TraA [Agrobacterium salinitolerans]
MAIAHFSVSIVSRGDGRSAVLSAAYRHCARMEYEREARTIDYTRKQGLLHEEFVLPADAPKWARMLIADR
SIAGASEAFWNKVEAFEKRSDAQFAKDLTIALPVELTPEQNIALVREFVEKHILAKGMVADWVYHDNAGN
PHIHLMTTLRPLTVDGFGAKKVAVTGEDGQPLRNKAGKIVYELWAGGTDDFNAFRDAWFERLNHHLALNG
ISLRVDGRSYEKQGIDLEPTIHLGVGAKAIERKAEKQGVRPELERIELNDARRAENTRRILRRPEIVLDL
ITRERSVFDERDVAKVLHRYVDDPAVFQQLMLSIILNPQVLRLQRDTIDFATAKKVPARYSTREMIRLEA
TMAGQAMWLSGRETHDVDDKVLAATFARHARLSDEQKAAIERIAGSARIAAVVGRAGAGKTTMMKAAREA
WEVAGYRVVGGALAGKAAEGLEKEAGIASRTLASWELRWQQGRDTLDARTVFVMDEAGMVASKQMAGFIE
AVVRSGAKIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREEWMRHASLDLARGNIEQGLLAYR
TRGHVVGESLKSEAVESLISAWNHDYDPAKSMLMLAHLRRDVRMLNMMAREKLVERGIIGEGHAFKTADG
TRQFDAGDQIVFLKNEGSLGVKNGMIGRVIEAAPNRMTVTVGEGDQRRQVTVESRFYNNVDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYFGRRSFEMNGGLAKVLSRRNAKETTLDYERGTLY
REALRFAENRGLHIVQVARTLLRDRLDWTLRQKSKLTDLANRLRALGERLGLQQSPPAQSAKEAKPMVAG
ITRFENSVADTVSERLDADPALKKQWEEVSIRFRYVFADPETAFQAMNFDAVLADKDAARQALKRLEAEP
ASIGPLRGKTGILASKTEREARRVAEINVPALRRDLERYLQMRQNAAQRLQADEQALRQRVSIDIPALSP
EARAVLERVRDAIDRNDLPAALAYALSNRETKLEIDGFNKAVTERFGERTLLTNAAREPSGKLFDKLSEG
MQPQEKERLKEAWPVMRTAQQLAAHERTTETLRQVEELRLAQRQTPVVKQ
MAIAHFSVSIVSRGDGRSAVLSAAYRHCARMEYEREARTIDYTRKQGLLHEEFVLPADAPKWARMLIADR
SIAGASEAFWNKVEAFEKRSDAQFAKDLTIALPVELTPEQNIALVREFVEKHILAKGMVADWVYHDNAGN
PHIHLMTTLRPLTVDGFGAKKVAVTGEDGQPLRNKAGKIVYELWAGGTDDFNAFRDAWFERLNHHLALNG
ISLRVDGRSYEKQGIDLEPTIHLGVGAKAIERKAEKQGVRPELERIELNDARRAENTRRILRRPEIVLDL
ITRERSVFDERDVAKVLHRYVDDPAVFQQLMLSIILNPQVLRLQRDTIDFATAKKVPARYSTREMIRLEA
TMAGQAMWLSGRETHDVDDKVLAATFARHARLSDEQKAAIERIAGSARIAAVVGRAGAGKTTMMKAAREA
WEVAGYRVVGGALAGKAAEGLEKEAGIASRTLASWELRWQQGRDTLDARTVFVMDEAGMVASKQMAGFIE
AVVRSGAKIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREEWMRHASLDLARGNIEQGLLAYR
TRGHVVGESLKSEAVESLISAWNHDYDPAKSMLMLAHLRRDVRMLNMMAREKLVERGIIGEGHAFKTADG
TRQFDAGDQIVFLKNEGSLGVKNGMIGRVIEAAPNRMTVTVGEGDQRRQVTVESRFYNNVDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYFGRRSFEMNGGLAKVLSRRNAKETTLDYERGTLY
REALRFAENRGLHIVQVARTLLRDRLDWTLRQKSKLTDLANRLRALGERLGLQQSPPAQSAKEAKPMVAG
ITRFENSVADTVSERLDADPALKKQWEEVSIRFRYVFADPETAFQAMNFDAVLADKDAARQALKRLEAEP
ASIGPLRGKTGILASKTEREARRVAEINVPALRRDLERYLQMRQNAAQRLQADEQALRQRVSIDIPALSP
EARAVLERVRDAIDRNDLPAALAYALSNRETKLEIDGFNKAVTERFGERTLLTNAAREPSGKLFDKLSEG
MQPQEKERLKEAWPVMRTAQQLAAHERTTETLRQVEELRLAQRQTPVVKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 10444 | GenBank | WP_217005262 |
| Name | traG_KHC17_RS24865_pDG3-1c |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 69017.99 Da Isoelectric Point: 9.0882
>WP_217005262.1 Ti-type conjugative transfer system protein TraG [Agrobacterium salinitolerans]
MTVNRLLLATLPAAFMIATMLAISGIETWLSGFGASPQAKLMLGRIGLALPYAAAAAIGVITLFATAGAA
NIKTAGLSVLAGASLVCIAAIAREGWRLFALADRVPRGQSVFAYTDPSAMLGACAAVFTGIFALRVAIRG
NATFAKAAPRRISGKRAVHGEADWMKMGDAARLFPDAGGIVIGERYRVDKDSMAAMPFRAGDAQSWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSSEVAPMVIDHRRKSGRNVIVLD
PATPAIGFNALDWIGRHGNTKEEDIVAVATWIMTDNARAASARDDFFRASAMQLLTALIADVCLSGHTNE
KDQTLRRVRSNLSEPEPKLRERLTRIYEQSESDFVKENVAVFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGDSFSTDDLAGGDADLFIALDLKVLEAHPGLARVVIGSLLNAIYNRNGEIEGRTLFLLDEVARLGY
LRILETARDAGRKYGITLTMIFQSIGQMREAYGGRDATSKWFESASWISFAAINDPDTADYISKRCGDTT
VEVDQTNRSTGMRGSSRSRSKQLSRRPLILPHEVLRMRADEQIVFTSGNPPLRCGRAIWFRRDDMKACVG
QNRFHREGE
MTVNRLLLATLPAAFMIATMLAISGIETWLSGFGASPQAKLMLGRIGLALPYAAAAAIGVITLFATAGAA
NIKTAGLSVLAGASLVCIAAIAREGWRLFALADRVPRGQSVFAYTDPSAMLGACAAVFTGIFALRVAIRG
NATFAKAAPRRISGKRAVHGEADWMKMGDAARLFPDAGGIVIGERYRVDKDSMAAMPFRAGDAQSWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSSEVAPMVIDHRRKSGRNVIVLD
PATPAIGFNALDWIGRHGNTKEEDIVAVATWIMTDNARAASARDDFFRASAMQLLTALIADVCLSGHTNE
KDQTLRRVRSNLSEPEPKLRERLTRIYEQSESDFVKENVAVFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGDSFSTDDLAGGDADLFIALDLKVLEAHPGLARVVIGSLLNAIYNRNGEIEGRTLFLLDEVARLGY
LRILETARDAGRKYGITLTMIFQSIGQMREAYGGRDATSKWFESASWISFAAINDPDTADYISKRCGDTT
VEVDQTNRSTGMRGSSRSRSKQLSRRPLILPHEVLRMRADEQIVFTSGNPPLRCGRAIWFRRDDMKACVG
QNRFHREGE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 10445 | GenBank | WP_217005158 |
| Name | t4cp2_KHC17_RS25170_pDG3-1c |
UniProt ID | _ |
| Length | 822 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 822 a.a. Molecular weight: 91445.13 Da Isoelectric Point: 5.7989
>WP_217005158.1 conjugal transfer protein TrbE [Agrobacterium salinitolerans]
MVALKSFRHSGPSFADLVPYAGLVNNGVILLKDGSLMAGWYFGGPDSESSTDAERNEVSRQINAILSRLG
SGWMIQVEAVRVPTDDYPADTDCHFPDPVTRAIDAERRAHFQKERGHFESRHALILTWRAPEARRSGLTR
YVYSDTASRSATYADKALESFLTSIREVEQYLANVVSIRRMMTRETSERGGSRVARYDELFQFIRFCITG
ENHPVRLPEIPMYLDWLVTAELQHGLTPLVENRFLGVVAIDGLPAESWPGILNALDLMPLTYRWSSRFVF
LDAEEARAKLERTRKKWQQKVRPFFDQLFQTQSRSADQDALTMVAETEDAIAEASSQLVAYGYYTPAIVI
FDEQQARLQEKCEAIRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLINTRNLADLIPLNSVWS
GSPTAPCPFYPPGSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYAGAQVFA
FDKGGSMLPLTLGLDGDHYQIGGDIGPSAEREGRALAFCPLAELSTDGDRAWAAEWIETLVALQGVTITP
DYRNAISRQIGLMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLALGAFQCFEIEEL
MNMGERNLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATALPKREYYVVSPDGRRLFDM
ALGPVALSFVGASGKEDLKRIRALHSEHGAAWPCPWLQQRGIAHADTLFPAQ
MVALKSFRHSGPSFADLVPYAGLVNNGVILLKDGSLMAGWYFGGPDSESSTDAERNEVSRQINAILSRLG
SGWMIQVEAVRVPTDDYPADTDCHFPDPVTRAIDAERRAHFQKERGHFESRHALILTWRAPEARRSGLTR
YVYSDTASRSATYADKALESFLTSIREVEQYLANVVSIRRMMTRETSERGGSRVARYDELFQFIRFCITG
ENHPVRLPEIPMYLDWLVTAELQHGLTPLVENRFLGVVAIDGLPAESWPGILNALDLMPLTYRWSSRFVF
LDAEEARAKLERTRKKWQQKVRPFFDQLFQTQSRSADQDALTMVAETEDAIAEASSQLVAYGYYTPAIVI
FDEQQARLQEKCEAIRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLINTRNLADLIPLNSVWS
GSPTAPCPFYPPGSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYAGAQVFA
FDKGGSMLPLTLGLDGDHYQIGGDIGPSAEREGRALAFCPLAELSTDGDRAWAAEWIETLVALQGVTITP
DYRNAISRQIGLMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLALGAFQCFEIEEL
MNMGERNLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATALPKREYYVVSPDGRRLFDM
ALGPVALSFVGASGKEDLKRIRALHSEHGAAWPCPWLQQRGIAHADTLFPAQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 93298..102997
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| KHC17_RS25115 (KHC17_25115) | 88554..89588 | + | 1035 | WP_217005137 | hydroxyacid dehydrogenase | - |
| KHC17_RS25120 (KHC17_25120) | 89585..90391 | + | 807 | WP_217005139 | sugar phosphate isomerase/epimerase | - |
| KHC17_RS25125 (KHC17_25125) | 90382..91629 | + | 1248 | WP_217005141 | glucarate dehydratase family protein | - |
| KHC17_RS25130 (KHC17_25130) | 91935..93194 | + | 1260 | WP_217005144 | MFS transporter | - |
| KHC17_RS25135 (KHC17_25135) | 93298..94626 | - | 1329 | WP_217005146 | IncP-type conjugal transfer protein TrbI | virB10 |
| KHC17_RS25140 (KHC17_25140) | 94639..95100 | - | 462 | WP_217005345 | conjugal transfer protein TrbH | - |
| KHC17_RS25145 (KHC17_25145) | 95100..95954 | - | 855 | WP_217005148 | P-type conjugative transfer protein TrbG | virB9 |
| KHC17_RS25150 (KHC17_25150) | 95972..96649 | - | 678 | WP_217005150 | conjugal transfer protein TrbF | virB8 |
| KHC17_RS25155 (KHC17_25155) | 96667..97860 | - | 1194 | WP_217005152 | P-type conjugative transfer protein TrbL | virB6 |
| KHC17_RS25160 (KHC17_25160) | 97854..98084 | - | 231 | WP_217005154 | entry exclusion protein TrbK | - |
| KHC17_RS25165 (KHC17_25165) | 98081..98890 | - | 810 | WP_217005156 | P-type conjugative transfer protein TrbJ | virB5 |
| KHC17_RS25170 (KHC17_25170) | 98862..101330 | - | 2469 | WP_217005158 | conjugal transfer protein TrbE | virb4 |
| KHC17_RS25175 (KHC17_25175) | 101340..101639 | - | 300 | WP_217005161 | conjugal transfer protein TrbD | virB3 |
| KHC17_RS25180 (KHC17_25180) | 101632..102036 | - | 405 | WP_217005163 | conjugal transfer pilin TrbC | virB2 |
| KHC17_RS25185 (KHC17_25185) | 102026..102997 | - | 972 | WP_217005165 | P-type conjugative transfer ATPase TrbB | virB11 |
| KHC17_RS30180 (KHC17_25190) | 102994..103643 | - | 650 | Protein_96 | acyl-homoserine-lactone synthase TraI | - |
| KHC17_RS25195 (KHC17_25195) | 103990..105207 | + | 1218 | WP_217005167 | plasmid partitioning protein RepA | - |
| KHC17_RS25200 (KHC17_25200) | 105430..106443 | + | 1014 | WP_160872130 | plasmid partitioning protein RepB | - |
| KHC17_RS25205 (KHC17_25205) | 106653..107987 | + | 1335 | WP_217005169 | plasmid replication protein RepC | - |
Host bacterium
| ID | 14460 | GenBank | NZ_CP074396 |
| Plasmid name | pDG3-1c | Incompatibility group | - |
| Plasmid size | 141550 bp | Coordinate of oriT [Strand] | 37246..37290 [+] |
| Host baterium | Agrobacterium salinitolerans strain DG3-1 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |