Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114017 |
| Name | oriT_KHPS1|unnamed |
| Organism | Pseudomonas sp. KHPS1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP100552 (48087..48194 [-], 108 nt) |
| oriT length | 108 nt |
| IRs (inverted repeats) | 70..75, 78..83 (ACCCGC..GCGGGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 108 nt
>oriT_KHPS1|unnamed
CCGGCGCCTCGCAGAGCAAGACACCCCCTTGAGCGCGTAGCGCGAATGTGGGGGAAGTGAAGAAGGCCGACCCGCTTGCGGGTGGGCCTACTTCACCTGTCTTGCCCG
CCGGCGCCTCGCAGAGCAAGACACCCCCTTGAGCGCGTAGCGCGAATGTGGGGGAAGTGAAGAAGGCCGACCCGCTTGCGGGTGGGCCTACTTCACCTGTCTTGCCCG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file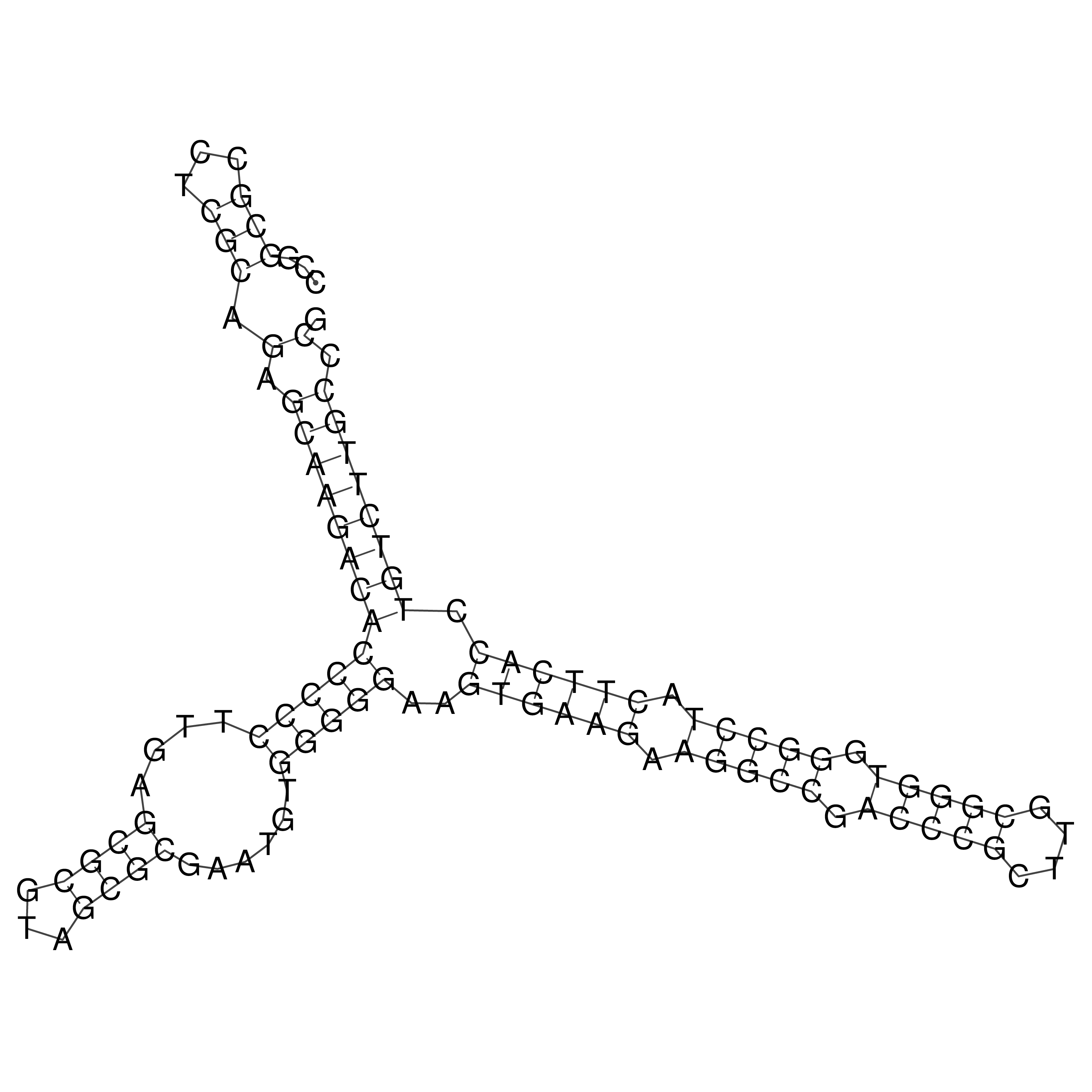
Relaxase
| ID | 9006 | GenBank | WP_254300308 |
| Name | traI_NLY39_RS23760_KHPS1|unnamed |
UniProt ID | _ |
| Length | 803 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 803 a.a. Molecular weight: 87575.28 Da Isoelectric Point: 10.7517
>WP_254300308.1 TraI/MobA(P) family conjugative relaxase [Pseudomonas sp. KHPS1]
MISKRIPMTSAKKSSFAGLVNYITNSQGISERVGEVRITNCQSEDLRWAVGEVLGTQQQNTRAEGDKTYH
LLISFAPGEKPSAEVLRDVEDRICAAMGYGEHQRVSAVHNDTDVLHIHVAINKIHPVRHTMHEPYRDFKT
RSDICAKLEIEHGLERVNHAGKKPYSESRANDMERHAGVESLLSWVRKECLEQMQGASSWKQLHQVLHEH
GLELREKGNGFVIQSQDGTMVKASTVARELSKAKLEGRLGAYEVRPSVGVQSLPAGAIPKKPGIGRVGRK
PPPLSRNRLRSLGSLQSLELDSGKRYEKRPIGNSDTTELYARYRDDQAAVRHTRTKELALERERRDRALD
YAKKESALKRASIKLMVTPGITKKLLYASAHQTLQKQLAKIRADHRKAVERINTTQKARQWADWLQAKAK
DGDQEALTALRAREGARGLRGDAIAGKNKPKATTVVRAAQEHITKEGTVIYRAGASAIRDDGSKLQLSRG
TDFDGIETALRMAVARYGEQITVTGSQQFKEQVAQTAALRGLPIKFDDPALEQRRQVLQQAIEKERSNVG
TDRGRAAGAGPGSDGIDTARRGRSGAGRNTTGATSGRPDGVRGYATGTGVPGRAGAGLGADGGRIAVRTT
GGTAKPGIARVGRKPPPESQNRLRTLSQLGVVQLTNGGESLLPSHVPGRVEQQGAATDNGLRWNVSRGGI
EGLPPSIDGIKAARQYIAEREDKRAKGFDIPKHRLYNAADEGMAAFAGVRSVEGQSMALLKRGDEVLVLP
INEATARRMKRMKLGEPVTTTAKGAIKSKGRSR
MISKRIPMTSAKKSSFAGLVNYITNSQGISERVGEVRITNCQSEDLRWAVGEVLGTQQQNTRAEGDKTYH
LLISFAPGEKPSAEVLRDVEDRICAAMGYGEHQRVSAVHNDTDVLHIHVAINKIHPVRHTMHEPYRDFKT
RSDICAKLEIEHGLERVNHAGKKPYSESRANDMERHAGVESLLSWVRKECLEQMQGASSWKQLHQVLHEH
GLELREKGNGFVIQSQDGTMVKASTVARELSKAKLEGRLGAYEVRPSVGVQSLPAGAIPKKPGIGRVGRK
PPPLSRNRLRSLGSLQSLELDSGKRYEKRPIGNSDTTELYARYRDDQAAVRHTRTKELALERERRDRALD
YAKKESALKRASIKLMVTPGITKKLLYASAHQTLQKQLAKIRADHRKAVERINTTQKARQWADWLQAKAK
DGDQEALTALRAREGARGLRGDAIAGKNKPKATTVVRAAQEHITKEGTVIYRAGASAIRDDGSKLQLSRG
TDFDGIETALRMAVARYGEQITVTGSQQFKEQVAQTAALRGLPIKFDDPALEQRRQVLQQAIEKERSNVG
TDRGRAAGAGPGSDGIDTARRGRSGAGRNTTGATSGRPDGVRGYATGTGVPGRAGAGLGADGGRIAVRTT
GGTAKPGIARVGRKPPPESQNRLRTLSQLGVVQLTNGGESLLPSHVPGRVEQQGAATDNGLRWNVSRGGI
EGLPPSIDGIKAARQYIAEREDKRAKGFDIPKHRLYNAADEGMAAFAGVRSVEGQSMALLKRGDEVLVLP
INEATARRMKRMKLGEPVTTTAKGAIKSKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 10438 | GenBank | WP_254300388 |
| Name | t4cp2_NLY39_RS23635_KHPS1|unnamed |
UniProt ID | _ |
| Length | 852 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 852 a.a. Molecular weight: 94657.92 Da Isoelectric Point: 6.4722
>WP_254300388.1 VirB4 family type IV secretion/conjugal transfer ATPase [Pseudomonas sp. KHPS1]
MIEGIAYAIAALGIVLVLILVQRLMTVDAELKLKKRRAKDAGLADLLIHAAVVDDGVIVGKNGSLMASWL
YKGDDNASSTEEQRELVSHRINQAFVGLGNGWMIHVDAVRRAAPNYSERGVSRFPDPLTAAIDEERRRLF
EGLGTMFEGYFVLTLTYFPPMLAQQKFVELMFDDEATPPDHKARTRNLIDQFKRDCASIEGSLSSVLKMS
RLRGNKVTLEDGRTVTHDDMLRWLQFCITGVSHPVMLPSNPMYLDAVLGGKELWGGVVPKVGRNFIQAVA
IEGFPLESAPGVLSALAEQPCEYRWSSRYIFMDAHEATKHLEKFRKKWKQKVRGFFDQVFNTNSGAIDQD
AMSMVQDAADAIAEVNSGLVSQGYFTSVVILMDENRERLRESAIKLEKLLERLGFAARIESINTMEAYLG
SIPGHGVENVRRPLIHTMNLADLLPTSTIWTGSNAAPCPLYPPLSPALMHCVTNGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLAILAAQLRRYEGMSIFAFDKGMSMYPLAAGINAATRGTSGLHFTVAADDEQLAFC
PLQFLETKADRAWAMEWIDTILALNGLNTSTEQRNHIGEAILTMHKDKSRTLSEFSINVQDQAIRSALAQ
YTVDGSMGHLLDAEEDGLALSDFTVFEIEELMNLGDKYALPVLLYLFRRIERALKGQPAVIILDEAWLML
GHPVFREKIKEWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTAALYRRMG
LNNRQIEILASAIPKRQYYYVSENGRRLYDLALGPLALAFVGSSDKESVAAIKTLQAKFGDRWVDEWLAA
RGLRINDYGVAA
MIEGIAYAIAALGIVLVLILVQRLMTVDAELKLKKRRAKDAGLADLLIHAAVVDDGVIVGKNGSLMASWL
YKGDDNASSTEEQRELVSHRINQAFVGLGNGWMIHVDAVRRAAPNYSERGVSRFPDPLTAAIDEERRRLF
EGLGTMFEGYFVLTLTYFPPMLAQQKFVELMFDDEATPPDHKARTRNLIDQFKRDCASIEGSLSSVLKMS
RLRGNKVTLEDGRTVTHDDMLRWLQFCITGVSHPVMLPSNPMYLDAVLGGKELWGGVVPKVGRNFIQAVA
IEGFPLESAPGVLSALAEQPCEYRWSSRYIFMDAHEATKHLEKFRKKWKQKVRGFFDQVFNTNSGAIDQD
AMSMVQDAADAIAEVNSGLVSQGYFTSVVILMDENRERLRESAIKLEKLLERLGFAARIESINTMEAYLG
SIPGHGVENVRRPLIHTMNLADLLPTSTIWTGSNAAPCPLYPPLSPALMHCVTNGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLAILAAQLRRYEGMSIFAFDKGMSMYPLAAGINAATRGTSGLHFTVAADDEQLAFC
PLQFLETKADRAWAMEWIDTILALNGLNTSTEQRNHIGEAILTMHKDKSRTLSEFSINVQDQAIRSALAQ
YTVDGSMGHLLDAEEDGLALSDFTVFEIEELMNLGDKYALPVLLYLFRRIERALKGQPAVIILDEAWLML
GHPVFREKIKEWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTAALYRRMG
LNNRQIEILASAIPKRQYYYVSENGRRLYDLALGPLALAFVGSSDKESVAAIKTLQAKFGDRWVDEWLAA
RGLRINDYGVAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 10439 | GenBank | WP_254300310 |
| Name | t4cp2_NLY39_RS23765_KHPS1|unnamed |
UniProt ID | _ |
| Length | 638 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 638 a.a. Molecular weight: 70246.51 Da Isoelectric Point: 8.5951
>WP_254300310.1 type IV secretory system conjugative DNA transfer family protein [Pseudomonas sp. KHPS1]
MISKDKFNNAVGPQVRAKKPGVGKIIPALSLLSIGVGLAASTQYFAYDFKYQDVLGPHYNHIYAPWSILT
WANKWYGTYPDAFMRAGSIGVLATGVGLLGLVITKMIQANSAKANEYLHGSARWANKKDIQASGLLPRSR
TLLEVVTGKEAPSSTGVYVGAWVDDQGVQHYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASTVVTD
LKGELWALTSGWRQKHARNKVLRFEPATTAGSVRWNPLDEIRIGTEYEVGDVQNLATLIVDPDGKGLDSH
WQKTSQALLVGLIMHVLYKSKNEGTPATLPEVDRILADPDRDLGELWMEMTTYSHSDGRNHPVVGSAARD
MMDRPEEEAGSVLSTAKSYLALYRDPVVARNVSASEFKVKDLMNHDNPVSLFIVTQPNDKARLRPLVRVL
INMIVRLLADKMDFDKGRPVAHYNHRLLMMLDEFPSLGKLEILQESLAFVAGYGIKCYLICQDINQLKSR
ETGYGPDETITSNCHVQNAYPPNRVETAEHLSKLTGQTTVVKEQITTSGRRTSAMLGQVSRTMQEVQRPL
LTPDECLRMPGPIKTASGDIAEAGDMVVYVAGYPAIYGKQPLYFKDPVFQARAAIPAPKVSDKLRVVQEQ
AEAEAITL
MISKDKFNNAVGPQVRAKKPGVGKIIPALSLLSIGVGLAASTQYFAYDFKYQDVLGPHYNHIYAPWSILT
WANKWYGTYPDAFMRAGSIGVLATGVGLLGLVITKMIQANSAKANEYLHGSARWANKKDIQASGLLPRSR
TLLEVVTGKEAPSSTGVYVGAWVDDQGVQHYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASTVVTD
LKGELWALTSGWRQKHARNKVLRFEPATTAGSVRWNPLDEIRIGTEYEVGDVQNLATLIVDPDGKGLDSH
WQKTSQALLVGLIMHVLYKSKNEGTPATLPEVDRILADPDRDLGELWMEMTTYSHSDGRNHPVVGSAARD
MMDRPEEEAGSVLSTAKSYLALYRDPVVARNVSASEFKVKDLMNHDNPVSLFIVTQPNDKARLRPLVRVL
INMIVRLLADKMDFDKGRPVAHYNHRLLMMLDEFPSLGKLEILQESLAFVAGYGIKCYLICQDINQLKSR
ETGYGPDETITSNCHVQNAYPPNRVETAEHLSKLTGQTTVVKEQITTSGRRTSAMLGQVSRTMQEVQRPL
LTPDECLRMPGPIKTASGDIAEAGDMVVYVAGYPAIYGKQPLYFKDPVFQARAAIPAPKVSDKLRVVQEQ
AEAEAITL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 23744..34370
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| NLY39_RS23560 (NLY39_23560) | 18951..19889 | + | 939 | WP_254300365 | LysR family transcriptional regulator | - |
| NLY39_RS23565 (NLY39_23565) | 19987..20652 | + | 666 | WP_254300366 | lipopolysaccharide assembly protein LapB | - |
| NLY39_RS23570 (NLY39_23570) | 21097..21246 | + | 150 | Protein_27 | AAA family ATPase | - |
| NLY39_RS23575 (NLY39_23575) | 21233..21475 | + | 243 | WP_254300368 | hypothetical protein | - |
| NLY39_RS23580 (NLY39_23580) | 21578..22303 | - | 726 | WP_254300370 | TraX family protein | - |
| NLY39_RS23585 (NLY39_23585) | 22307..22777 | - | 471 | WP_254300372 | hypothetical protein | - |
| NLY39_RS23590 (NLY39_23590) | 22777..23406 | - | 630 | WP_254300374 | muramidase | - |
| NLY39_RS23595 (NLY39_23595) | 23416..23718 | - | 303 | WP_254300376 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| NLY39_RS23600 (NLY39_23600) | 23744..25489 | - | 1746 | WP_254300379 | P-type conjugative transfer protein TrbL | virB6 |
| NLY39_RS23605 (NLY39_23605) | 25511..25747 | - | 237 | WP_254300381 | entry exclusion lipoprotein TrbK | - |
| NLY39_RS23610 (NLY39_23610) | 25762..26553 | - | 792 | WP_254300382 | P-type conjugative transfer protein TrbJ | virB5 |
| NLY39_RS23615 (NLY39_23615) | 26576..27970 | - | 1395 | WP_254300404 | TrbI/VirB10 family protein | virB10 |
| NLY39_RS23620 (NLY39_23620) | 27999..28466 | - | 468 | WP_254300384 | conjugal transfer protein TrbH | - |
| NLY39_RS23625 (NLY39_23625) | 28470..29360 | - | 891 | WP_031943183 | P-type conjugative transfer protein TrbG | virB9 |
| NLY39_RS23630 (NLY39_23630) | 29372..30097 | - | 726 | WP_254300386 | conjugal transfer protein TrbF | virB8 |
| NLY39_RS23635 (NLY39_23635) | 30094..32652 | - | 2559 | WP_254300388 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| NLY39_RS23640 (NLY39_23640) | 32649..32960 | - | 312 | WP_254300390 | conjugal transfer protein TrbD | virB3 |
| NLY39_RS23645 (NLY39_23645) | 32963..33370 | - | 408 | WP_031943179 | conjugal transfer system pilin TrbC | virB2 |
| NLY39_RS23650 (NLY39_23650) | 33402..34370 | - | 969 | WP_254300392 | P-type conjugative transfer ATPase TrbB | virB11 |
| NLY39_RS23655 (NLY39_23655) | 34625..34996 | - | 372 | WP_046935729 | transcriptional regulator | - |
| NLY39_RS23660 (NLY39_23660) | 35106..35465 | + | 360 | WP_254300395 | single-stranded DNA-binding protein | - |
| NLY39_RS23665 (NLY39_23665) | 35510..36598 | + | 1089 | WP_254300397 | plasmid replication initiator TrfA | - |
| NLY39_RS23670 (NLY39_23670) | 36602..36814 | + | 213 | WP_073624929 | hypothetical protein | - |
| NLY39_RS23675 (NLY39_23675) | 38515..38937 | + | 423 | WP_254300399 | antirestriction protein | - |
Host bacterium
| ID | 14452 | GenBank | NZ_CP100552 |
| Plasmid name | KHPS1|unnamed | Incompatibility group | - |
| Plasmid size | 79836 bp | Coordinate of oriT [Strand] | 48087..48194 [-] |
| Host baterium | Pseudomonas sp. KHPS1 |
Cargo genes
| Drug resistance gene | qacE |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |