Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 114007 |
| Name | oriT_p19-0822-3296 |
| Organism | Shigella sonnei strain 19.0822.3296 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP049184 (37510..37598 [+], 89 nt) |
| oriT length | 89 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 89 nt
>oriT_p19-0822-3296
GGGGTGTCGGGGCGAAGCCCTGACCAGATGGTAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTACACATCCTGTCCCGTTTTTC
GGGGTGTCGGGGCGAAGCCCTGACCAGATGGTAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTACACATCCTGTCCCGTTTTTC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file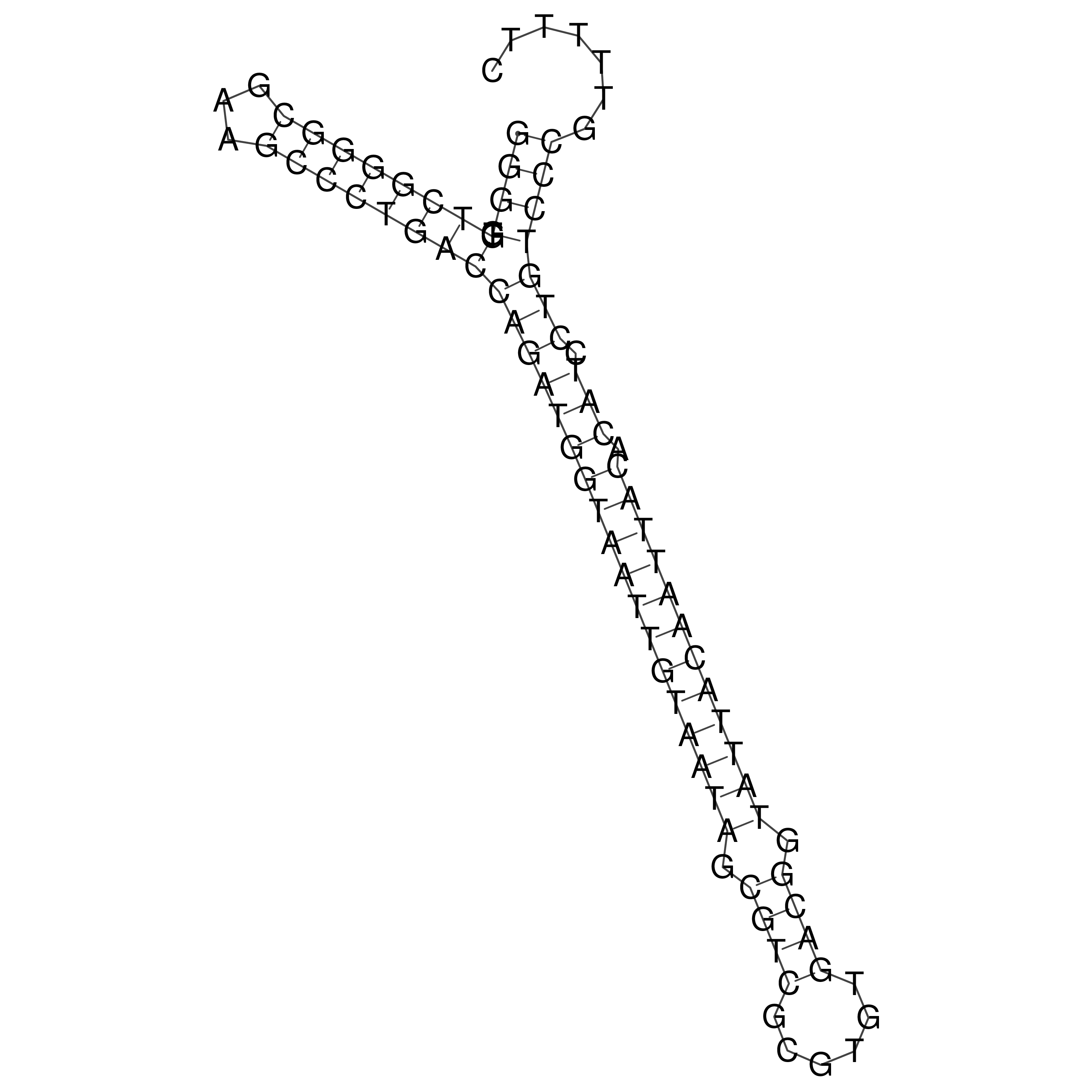
Relaxase
| ID | 8999 | GenBank | WP_011264053 |
| Name | nikB_G6O66_RS24360_p19-0822-3296 |
UniProt ID | A0A767TAZ8 |
| Length | 899 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 103882.34 Da Isoelectric Point: 7.4198
>WP_011264053.1 MULTISPECIES: IncI1-type relaxase NikB [Enterobacteriaceae]
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELNLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIAQQAHYAKDNTDPVFHYILSWQAHESPRPEQIYDSVR
HTLKSLGLGEHQYVSAVHTDTDNLHVHVAVNRVHPVTGYLNCLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVNDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELNLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIAQQAHYAKDNTDPVFHYILSWQAHESPRPEQIYDSVR
HTLKSLGLGEHQYVSAVHTDTDNLHVHVAVNRVHPVTGYLNCLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVNDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A767TAZ8 |
Auxiliary protein
| ID | 5246 | GenBank | WP_063119516 |
| Name | WP_063119516_p19-0822-3296 |
UniProt ID | _ |
| Length | 110 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 110 a.a. Molecular weight: 12585.55 Da Isoelectric Point: 10.6897
>WP_063119516.1 MULTISPECIES: IncI1-type relaxosome accessory protein NikA [Enterobacteriaceae]
MSDSAVKKKSEVRQKTVVRTLRFSPVEDETIRKKAEDSGLTVSAYIRNAALNKRINSRTDDAFLKELMRL
GRMQKHLFVQGKRTGDKEYAEVLVAITELTNTLRKQLMEG
MSDSAVKKKSEVRQKTVVRTLRFSPVEDETIRKKAEDSGLTVSAYIRNAALNKRINSRTDDAFLKELMRL
GRMQKHLFVQGKRTGDKEYAEVLVAITELTNTLRKQLMEG
Protein domains
No domain identified.
Protein structure
No available structure.
T4CP
| ID | 10434 | GenBank | WP_011264051 |
| Name | trbC_G6O66_RS24355_p19-0822-3296 |
UniProt ID | _ |
| Length | 763 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 763 a.a. Molecular weight: 86954.10 Da Isoelectric Point: 6.7713
>WP_011264051.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Enterobacteriaceae]
MSEHRVNPELLHRTAWGNPVWNALQSLNIYGFCLVASLVASFIWPLALPACLLFTLITMLVFSLQRWRCP
LRMPMTLECADPSQDRMIKRSLFSFWPTLFQYEVILESPASGIFYVGYQRVRDIGRELWLSMDDLTRHIM
FFATTGGGKTETIFAWAINPLCWARGFTLVDGKAQNDTARTIWYLARRFGREDDVEVINFMNGGKSRSEI
ILSGEKTRPQSNTWNPFCYSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDLSLVRTPSAWTEEPRKQHAYLSGQFSE
TFSTFTEAFGDIFAEDSGDIDIRDSIHSDRILMVMIPALDTSAHTTSALGRMFITQKSMILARDLGYRLE
GTDSDALEVKKYKGRFPYLCFLDEVGAYYTDRIAVEATQVRSLDFALILMAQDQERIEGQTTATNTATLM
QNTGTKFAGRIVSEGSTARTLKSAAGEEARARMNNLQRQDGIFGESWIDSPQISILMESKINVQELIELH
PGEFFSIFRGETVPSASFFIPDDEKSCSSDPVVINRYISVDAPRLDRLRRLVPRTTQRRIPSPENVSAII
GVLTAKPSRKRRKIRTEPHTIVDTFQQRIAGRQAAMAMLEEYDTDINARESALWETAVNTLKTTTREERR
IRYITLNRPELPETKEENQISVRAERAGINLLTLPQDNNHPTGRPVNGFHHKKNNRPDWDGMY
MSEHRVNPELLHRTAWGNPVWNALQSLNIYGFCLVASLVASFIWPLALPACLLFTLITMLVFSLQRWRCP
LRMPMTLECADPSQDRMIKRSLFSFWPTLFQYEVILESPASGIFYVGYQRVRDIGRELWLSMDDLTRHIM
FFATTGGGKTETIFAWAINPLCWARGFTLVDGKAQNDTARTIWYLARRFGREDDVEVINFMNGGKSRSEI
ILSGEKTRPQSNTWNPFCYSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDLSLVRTPSAWTEEPRKQHAYLSGQFSE
TFSTFTEAFGDIFAEDSGDIDIRDSIHSDRILMVMIPALDTSAHTTSALGRMFITQKSMILARDLGYRLE
GTDSDALEVKKYKGRFPYLCFLDEVGAYYTDRIAVEATQVRSLDFALILMAQDQERIEGQTTATNTATLM
QNTGTKFAGRIVSEGSTARTLKSAAGEEARARMNNLQRQDGIFGESWIDSPQISILMESKINVQELIELH
PGEFFSIFRGETVPSASFFIPDDEKSCSSDPVVINRYISVDAPRLDRLRRLVPRTTQRRIPSPENVSAII
GVLTAKPSRKRRKIRTEPHTIVDTFQQRIAGRQAAMAMLEEYDTDINARESALWETAVNTLKTTTREERR
IRYITLNRPELPETKEENQISVRAERAGINLLTLPQDNNHPTGRPVNGFHHKKNNRPDWDGMY
Protein domains
No domain identified.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1306..32132
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| G6O66_RS24190 | 1..1155 | + | 1155 | WP_001139958 | tyrosine-type recombinase/integrase | - |
| G6O66_RS24195 (G6O66_24145) | 1306..2130 | + | 825 | WP_001545755 | conjugal transfer protein TraE | traE |
| G6O66_RS24200 (G6O66_24150) | 2216..3418 | + | 1203 | WP_000976353 | conjugal transfer protein TraF | - |
| G6O66_RS24205 (G6O66_24155) | 3478..4062 | + | 585 | WP_000977522 | histidine phosphatase family protein | - |
| G6O66_RS24210 (G6O66_24160) | 4457..4915 | + | 459 | WP_001079808 | IncI1-type conjugal transfer lipoprotein TraH | - |
| G6O66_RS24215 (G6O66_24165) | 4912..5730 | + | 819 | WP_097763371 | IncI1-type conjugal transfer lipoprotein TraI | traI |
| G6O66_RS24220 (G6O66_24170) | 5727..6875 | + | 1149 | WP_001024972 | plasmid transfer ATPase TraJ | virB11 |
| G6O66_RS24225 (G6O66_24175) | 6872..7162 | + | 291 | WP_001314267 | hypothetical protein | traK |
| G6O66_RS24230 (G6O66_24180) | 7177..7728 | + | 552 | WP_000014586 | phospholipase D family protein | - |
| G6O66_RS24235 (G6O66_24185) | 7818..11582 | + | 3765 | WP_001141540 | LPD7 domain-containing protein | - |
| G6O66_RS24240 (G6O66_24190) | 11600..11947 | + | 348 | WP_001055900 | conjugal transfer protein | traL |
| G6O66_RS24245 (G6O66_24195) | 11944..12636 | + | 693 | WP_000138552 | DotI/IcmL family type IV secretion protein | traM |
| G6O66_RS24250 (G6O66_24200) | 12647..13630 | + | 984 | WP_001191879 | IncI1-type conjugal transfer protein TraN | traN |
| G6O66_RS24255 (G6O66_24205) | 13633..14922 | + | 1290 | WP_001271994 | conjugal transfer protein TraO | traO |
| G6O66_RS24260 (G6O66_24210) | 14922..15626 | + | 705 | WP_000801920 | IncI1-type conjugal transfer protein TraP | traP |
| G6O66_RS24265 (G6O66_24215) | 15626..16153 | + | 528 | WP_001055569 | conjugal transfer protein TraQ | traQ |
| G6O66_RS24270 (G6O66_24220) | 16204..16608 | + | 405 | WP_000086965 | IncI1-type conjugal transfer protein TraR | traR |
| G6O66_RS24275 (G6O66_24225) | 16672..16860 | + | 189 | WP_001277253 | putative conjugal transfer protein TraS | - |
| G6O66_RS24280 (G6O66_24230) | 16844..17644 | + | 801 | WP_107372244 | IncI1-type conjugal transfer protein TraT | traT |
| G6O66_RS24285 (G6O66_24235) | 17734..20778 | + | 3045 | WP_107372243 | IncI1-type conjugal transfer protein TraU | traU |
| G6O66_RS24290 (G6O66_24240) | 20778..21392 | + | 615 | WP_000337395 | IncI1-type conjugal transfer protein TraV | traV |
| G6O66_RS24295 (G6O66_24245) | 21359..22561 | + | 1203 | WP_107372242 | IncI1-type conjugal transfer protein TraW | traW |
| G6O66_RS24300 (G6O66_24250) | 22590..23174 | + | 585 | WP_001037987 | IncI1-type conjugal transfer protein TraX | - |
| G6O66_RS24305 (G6O66_24255) | 23271..25439 | + | 2169 | WP_001774191 | DotA/TraY family protein | traY |
| G6O66_RS24310 (G6O66_24260) | 25513..26163 | + | 651 | WP_001178506 | plasmid IncI1-type surface exclusion protein ExcA | - |
| G6O66_RS24315 (G6O66_24265) | 26235..26444 | - | 210 | WP_000062603 | HEAT repeat domain-containing protein | - |
| G6O66_RS24320 | 26810..26986 | + | 177 | WP_001054898 | hypothetical protein | - |
| G6O66_RS24795 | 27051..27146 | - | 96 | WP_000609148 | DinQ-like type I toxin DqlB | - |
| G6O66_RS24330 (G6O66_24275) | 27647..27898 | + | 252 | WP_001291964 | hypothetical protein | - |
| G6O66_RS24335 (G6O66_24280) | 27970..28122 | - | 153 | WP_001387489 | Hok/Gef family protein | - |
| G6O66_RS24340 (G6O66_24285) | 28749..29528 | + | 780 | WP_275450201 | protein FinQ | - |
| G6O66_RS24345 (G6O66_24290) | 29835..31043 | + | 1209 | WP_107372241 | IncI1-type conjugal transfer protein TrbA | trbA |
| G6O66_RS24350 (G6O66_24295) | 31062..32132 | + | 1071 | WP_000151590 | IncI1-type conjugal transfer protein TrbB | trbB |
| G6O66_RS24355 (G6O66_24300) | 32125..34416 | + | 2292 | WP_011264051 | F-type conjugative transfer protein TrbC | - |
Host bacterium
| ID | 14442 | GenBank | NZ_CP049184 |
| Plasmid name | p19-0822-3296 | Incompatibility group | IncI1 |
| Plasmid size | 87623 bp | Coordinate of oriT [Strand] | 37510..37598 [+] |
| Host baterium | Shigella sonnei strain 19.0822.3296 |
Cargo genes
| Drug resistance gene | blaCTX-M-3 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrVA2 |