Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 113971 |
| Name | oriT_pWBG731 |
| Organism | Staphylococcus aureus strain WBG10514 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_MH587574 (15709..15853 [+], 145 nt) |
| oriT length | 145 nt |
| IRs (inverted repeats) | 105..110, 114..119 (TTTAAA..TTTAAA) 33..39, 44..50 (TGTCACA..TGTGACA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 145 nt
>oriT_pWBG731
TGTCACAAAAATAAGACAAATTCTACATGTTGTGTCACAAAACTGTGACAAACACAATATATAGTGTCACAAAACTGTGACACTATAGTTTATTGTGGGGTCACTTTAAATTCTTTAAAAACCTTGAAATGGCTGGCTTTGCCAG
TGTCACAAAAATAAGACAAATTCTACATGTTGTGTCACAAAACTGTGACAAACACAATATATAGTGTCACAAAACTGTGACACTATAGTTTATTGTGGGGTCACTTTAAATTCTTTAAAAACCTTGAAATGGCTGGCTTTGCCAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file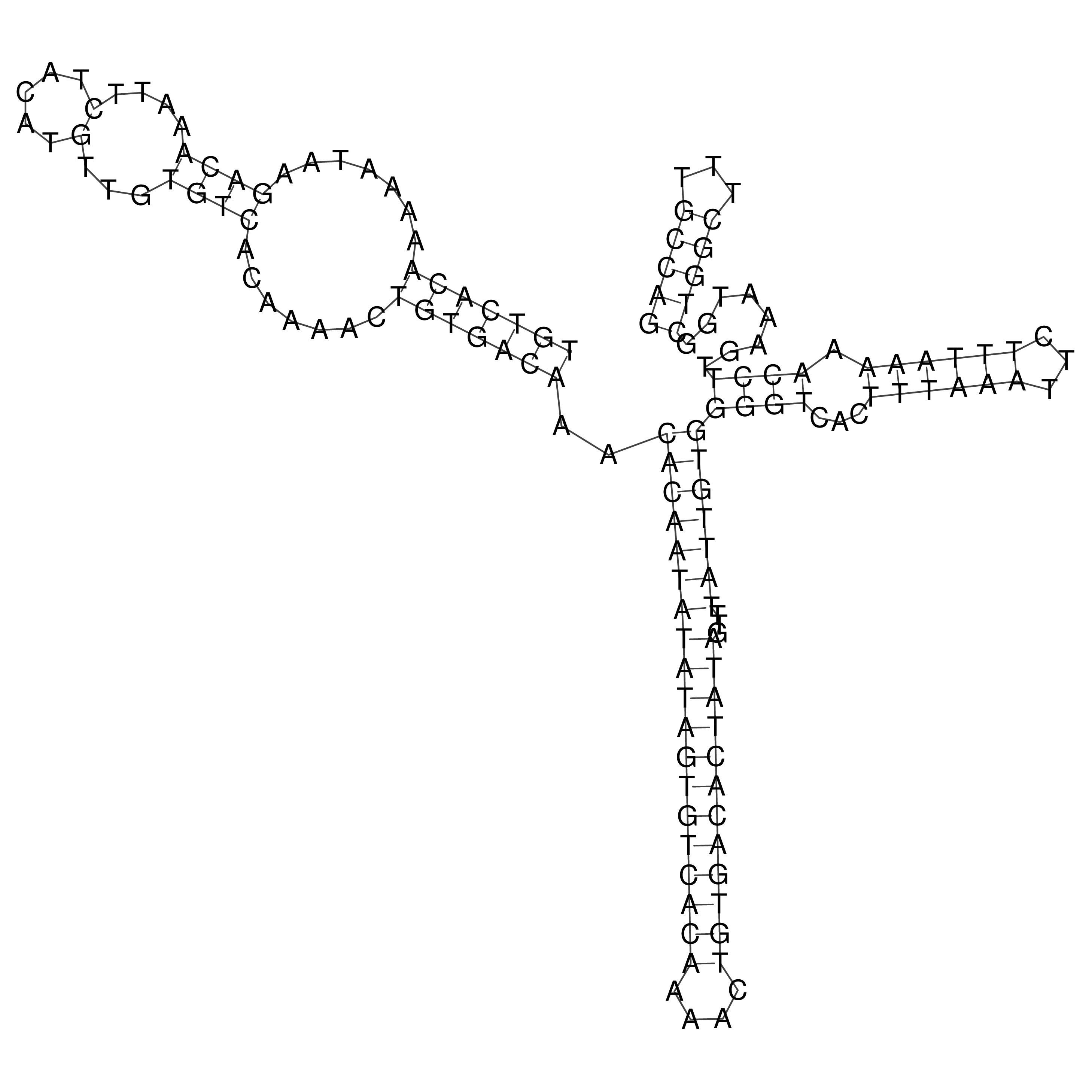
Relaxase
| ID | 8973 | GenBank | WP_223223918 |
| Name | mobP2_HTT44_RS00095_pWBG731 |
UniProt ID | _ |
| Length | 383 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 383 a.a. Molecular weight: 45796.08 Da Isoelectric Point: 9.9143
>WP_223223918.1 MULTISPECIES: MobP2 family relaxase [Staphylococcus]
MMPKIVTVSQFEQPNEFYEYSNYVNYMKRGEAQNNKDDFKYDVYAHYMFDEEKSQNMFNEHDNFMSKKAI
NKIKSDFSHAQKNKGMMWKDVISFDNSALESVGIYDSKTHTLDEQKIKEATRKMMKRFEKKEGLEDNLVW
SGAIHYNTDNIHVHVAAVEKVIKRTRGKRKPATLKHMKSTFANELFDIKGERQKINEFIRERIVKGIRDE
SEPCKDKGMKKQIKKVHKLLEDIPRNQWNYNNNILKYIRPEIDKITDIYIKNYHSKDFEVFKNDLKRQTN
LYKETYGENSNYGSYEKTKMKDLYARSGNTILKNIKELDQDLMKVKQQSTHSKSNATITKLKIGKCLNQA
LYRTKYALRSDYDKEKNIQQYYQEFDKQSYRER
MMPKIVTVSQFEQPNEFYEYSNYVNYMKRGEAQNNKDDFKYDVYAHYMFDEEKSQNMFNEHDNFMSKKAI
NKIKSDFSHAQKNKGMMWKDVISFDNSALESVGIYDSKTHTLDEQKIKEATRKMMKRFEKKEGLEDNLVW
SGAIHYNTDNIHVHVAAVEKVIKRTRGKRKPATLKHMKSTFANELFDIKGERQKINEFIRERIVKGIRDE
SEPCKDKGMKKQIKKVHKLLEDIPRNQWNYNNNILKYIRPEIDKITDIYIKNYHSKDFEVFKNDLKRQTN
LYKETYGENSNYGSYEKTKMKDLYARSGNTILKNIKELDQDLMKVKQQSTHSKSNATITKLKIGKCLNQA
LYRTKYALRSDYDKEKNIQQYYQEFDKQSYRER
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 10411 | GenBank | WP_042853329 |
| Name | t4cp2_HTT44_RS00045_pWBG731 |
UniProt ID | _ |
| Length | 924 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 924 a.a. Molecular weight: 106282.13 Da Isoelectric Point: 7.5736
>WP_042853329.1 VirD4-like conjugal transfer protein, CD1115 family [Staphylococcus aureus]
MRFGSKDKHGYRRKKLSFSQNKHLLAHPCILVSTLLLIMLLATAIANFLINILMTVFFGNHDIFTNMKQL
NTKYTDYIFNFRSDAVLLYIPIWGLVFIFFGKKVYQFRQRFKSLNNNEKASGRFATRKEITQQYKAIPQR
EHAFEGEGGLPVAMFGVYDFVTKTKQYLEQRKTAIREINGITDLTADEKLELRKDKLGKMNKEFATTFFS
KREFTFIDESPTNNLIVGTSRSGKGEIVVLSMIENYSRSSEQPSLIVNDMKGELFSASYKTLENRGYLVE
IFNLDQPMESTMSFNPLQQVIDEYMKGDLGEAQEMTKQITYTLTNNEGVEDNEWNLMAGALINAMILALC
EETLPKNPEKVTLYSVSNMLQTLSTKRFYKEIAIGNKVQLIEVSALDEYMERFPSHSPAKTQYATVQAAP
DKMRSSIIGTALKALQPFTTDTIAKLTSHTSFDINKLGFPSIIDGYATKDSTFELGVQVLRDGENGIEYE
EVEGIQIGVNEYGYWQYPFKTVLKVGDRIETIEKDGNNQVITSYFYVDHIDDKGQVTFNQKGELDNSDIT
IRKFNSFPKPIAVFMVVPDYNNANHGIASILVNQIVFEISKNSQLYTENQSTYRRVIFHLDEAGNMPPIP
NLSQKVNVSLSRGLRFNFFVQAFSQFKDKYGDAYDAIMDACQNKVYIMATQEQTLKDFSAMIGSQQITVH
SRSGEAGAIKSSITENQEERPLLRPDELARLQEGETVVVRNLKRQDKKRNKVMSYPIFNDGKYAMKYRYQ
YLSDLMDTSQSIIHFRPMLKARCEHRHLQLEATLVDWDKRIEEMQEGIDGQKEQENKNINDINQYKQSNE
EGNVAVRVKRLETLKEKVGDAIFDKITRRFERYLRTYAEQGKNVNTITQKKLEELVSADNEVKEEEKTKR
IEMIQKFIKEAKTT
MRFGSKDKHGYRRKKLSFSQNKHLLAHPCILVSTLLLIMLLATAIANFLINILMTVFFGNHDIFTNMKQL
NTKYTDYIFNFRSDAVLLYIPIWGLVFIFFGKKVYQFRQRFKSLNNNEKASGRFATRKEITQQYKAIPQR
EHAFEGEGGLPVAMFGVYDFVTKTKQYLEQRKTAIREINGITDLTADEKLELRKDKLGKMNKEFATTFFS
KREFTFIDESPTNNLIVGTSRSGKGEIVVLSMIENYSRSSEQPSLIVNDMKGELFSASYKTLENRGYLVE
IFNLDQPMESTMSFNPLQQVIDEYMKGDLGEAQEMTKQITYTLTNNEGVEDNEWNLMAGALINAMILALC
EETLPKNPEKVTLYSVSNMLQTLSTKRFYKEIAIGNKVQLIEVSALDEYMERFPSHSPAKTQYATVQAAP
DKMRSSIIGTALKALQPFTTDTIAKLTSHTSFDINKLGFPSIIDGYATKDSTFELGVQVLRDGENGIEYE
EVEGIQIGVNEYGYWQYPFKTVLKVGDRIETIEKDGNNQVITSYFYVDHIDDKGQVTFNQKGELDNSDIT
IRKFNSFPKPIAVFMVVPDYNNANHGIASILVNQIVFEISKNSQLYTENQSTYRRVIFHLDEAGNMPPIP
NLSQKVNVSLSRGLRFNFFVQAFSQFKDKYGDAYDAIMDACQNKVYIMATQEQTLKDFSAMIGSQQITVH
SRSGEAGAIKSSITENQEERPLLRPDELARLQEGETVVVRNLKRQDKKRNKVMSYPIFNDGKYAMKYRYQ
YLSDLMDTSQSIIHFRPMLKARCEHRHLQLEATLVDWDKRIEEMQEGIDGQKEQENKNINDINQYKQSNE
EGNVAVRVKRLETLKEKVGDAIFDKITRRFERYLRTYAEQGKNVNTITQKKLEELVSADNEVKEEEKTKR
IEMIQKFIKEAKTT
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 14406 | GenBank | NZ_MH587574 |
| Plasmid name | pWBG731 | Incompatibility group | - |
| Plasmid size | 72158 bp | Coordinate of oriT [Strand] | 15709..15853 [+] |
| Host baterium | Staphylococcus aureus strain WBG10514 |
Cargo genes
| Drug resistance gene | blaZ, mupA |
| Virulence gene | - |
| Metal resistance gene | cadC |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |