Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 113968 |
| Name | oriT_pCH11 |
| Organism | Staphylococcus aureus strain ch11 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_MH785227 (2817..3056 [+], 240 nt) |
| oriT length | 240 nt |
| IRs (inverted repeats) | 217..222, 232..237 (ATTTTA..TAAAAT) 171..178, 183..190 (CTATCATT..AATGATAG) 154..160, 164..170 (GTCTGGC..GCCAGAC) 87..92, 99..104 (TGTCAC..GTGACA) 55..61, 66..72 (TGTCACA..TGTGACA) 27..33, 44..50 (TTTTTTA..TAAAAAA) 37..42, 45..50 (TTTTTT..AAAAAA) 26..31, 45..50 (TTTTTT..AAAAAA) 1..7, 20..26 (AAGACAT..ATGTCTT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 240 nt
>oriT_pCH11
AAGACATTAGTGATAACTGATGTCTTTTTTTTATTGTTTTTTGTAAAAAAGTGCTGTCACAAAACTGTGACAAACACAATATATTGTGTCACAAAAGCGTGACATTTTATCGTAATGTGATATCACTTTGAATTAACTTAAAAGTCTTGGAATGTCTGGCTTTGCCAGACCTATCATTTTTGAATGATAGCAAAATTCCCTTATGCTCTTACGGAGATTTTAGAGTTAAATTAAAATTTC
AAGACATTAGTGATAACTGATGTCTTTTTTTTATTGTTTTTTGTAAAAAAGTGCTGTCACAAAACTGTGACAAACACAATATATTGTGTCACAAAAGCGTGACATTTTATCGTAATGTGATATCACTTTGAATTAACTTAAAAGTCTTGGAATGTCTGGCTTTGCCAGACCTATCATTTTTGAATGATAGCAAAATTCCCTTATGCTCTTACGGAGATTTTAGAGTTAAATTAAAATTTC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file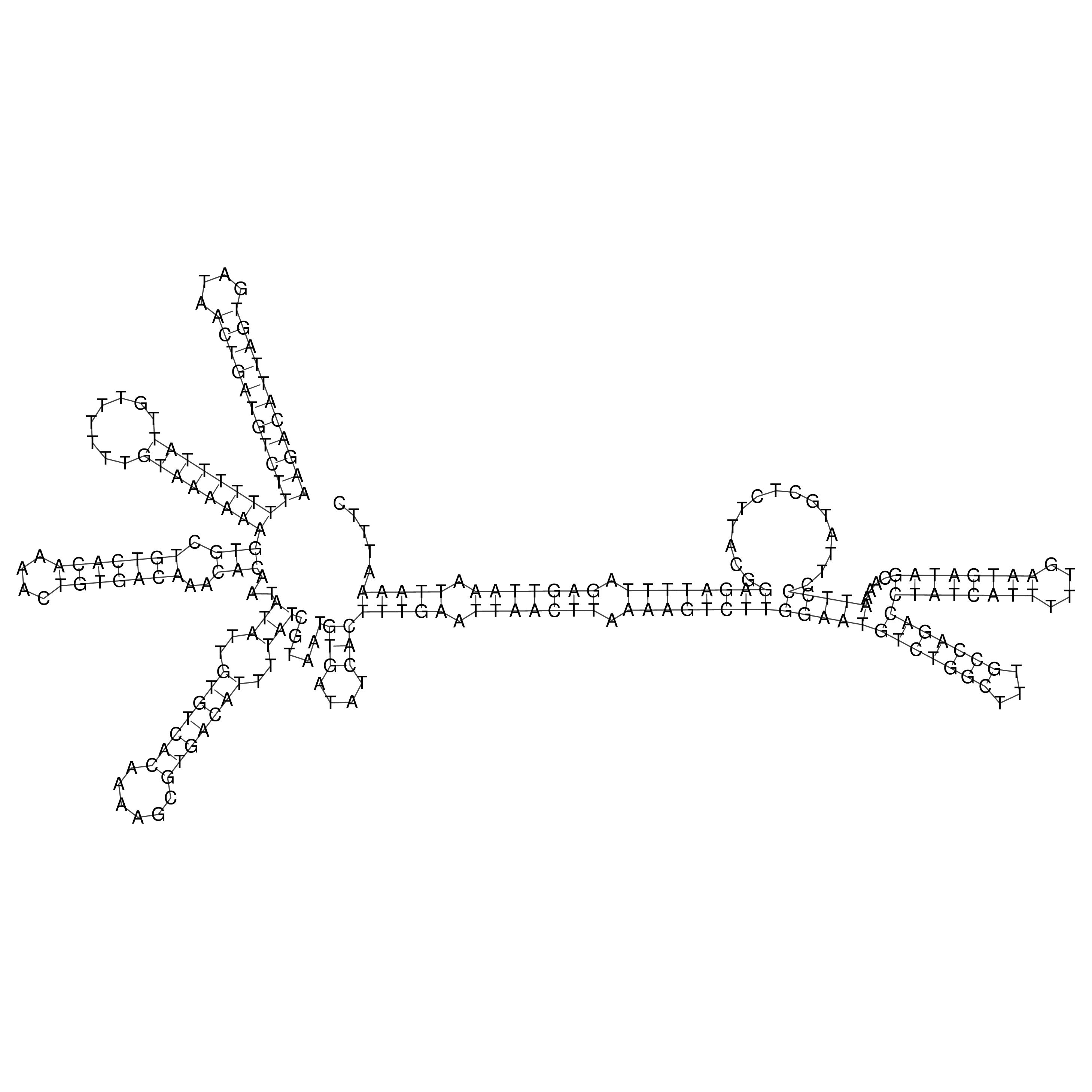
Host bacterium
| ID | 14403 | GenBank | NZ_MH785227 |
| Plasmid name | pCH11 | Incompatibility group | - |
| Plasmid size | 3259 bp | Coordinate of oriT [Strand] | 2817..3056 [+] |
| Host baterium | Staphylococcus aureus strain ch11 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |