Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 113921 |
| Name | oriT_pMCR_1410 |
| Organism | Kluyvera ascorbata strain WCH1410 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_KU922754 (6374..6426 [-], 53 nt) |
| oriT length | 53 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 53 nt
>oriT_pMCR_1410
CACACGATTGTAACATGACCGGAACGGTCTTGTGTACAATCGGTATCGTGCCT
CACACGATTGTAACATGACCGGAACGGTCTTGTGTACAATCGGTATCGTGCCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file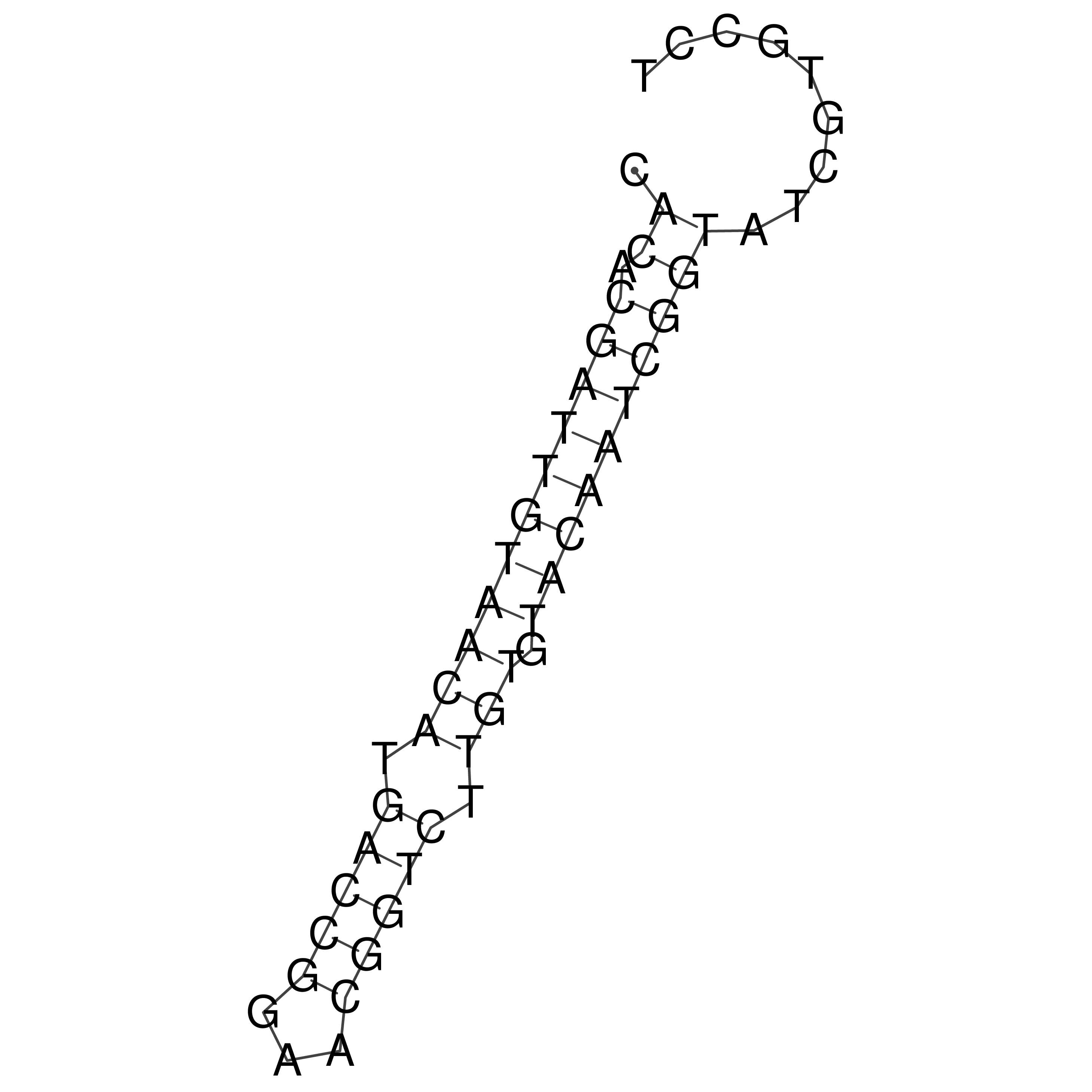
T4CP
| ID | 10377 | GenBank | WP_000338976 |
| Name | t4cp2_HTH65_RS00175_pMCR_1410 |
UniProt ID | _ |
| Length | 652 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 652 a.a. Molecular weight: 73346.95 Da Isoelectric Point: 9.3476
>WP_000338976.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Enterobacteriaceae]
MDAKKTGGLILFLLLLLVGVLIASNYLGGYTALRYSSVDMSLLKWDTFHSVISTFSGNPQYKKLVFMAWF
GFSVPLIFFAIFMLIVVIGIMPKKVIYGDARLATDMDLSKSGFFPDKKSPYKHPPILIGKMFKGRYKKQF
IYFAGQQFLILYAPTRSGKGVGIVIPNCVNYPGSMVILDIKLENWFLSAGFRQKELGQKCFLFAPAGYAE
TIDQAIKGQIRSHRWNPLDCVSRSDLLRETDLAKIAAILIPASDDPIWSDSARNLFVGLGLYLLDKERFH
LEQKAKGHNVPDVLVSISAILKTSVPDGGKDLAAWMGQEIENRSWISDKTKSFFFKFMSAPDRTRGSIET
NFSSPLSIFSNPITAEATNFSDFDIRDIRKKPMSIYLGLTPDALITHEKIVNLFFSLLVNENCRELPEHN
PDLKYQCLILLDEFTSMGKSEVIERAVGFTAGYNLRFMFILQNEGQGQKSDMYGQEGWTTFTENSAVVLY
YPPKSKNALAKKISEEIGVRDMKISKRSISSGGGKGGSSRTRNDDVIERPVLLPEEIVSLRDKKNKARNI
AIREIITSEFSRPFIANKIIWFEEPEFKRRVDIARNNHVDIPNLFTQEVMDEIAKIAEIYLPKAGGKKVM
VAGGNVITNPDLDNHDKTDVSE
MDAKKTGGLILFLLLLLVGVLIASNYLGGYTALRYSSVDMSLLKWDTFHSVISTFSGNPQYKKLVFMAWF
GFSVPLIFFAIFMLIVVIGIMPKKVIYGDARLATDMDLSKSGFFPDKKSPYKHPPILIGKMFKGRYKKQF
IYFAGQQFLILYAPTRSGKGVGIVIPNCVNYPGSMVILDIKLENWFLSAGFRQKELGQKCFLFAPAGYAE
TIDQAIKGQIRSHRWNPLDCVSRSDLLRETDLAKIAAILIPASDDPIWSDSARNLFVGLGLYLLDKERFH
LEQKAKGHNVPDVLVSISAILKTSVPDGGKDLAAWMGQEIENRSWISDKTKSFFFKFMSAPDRTRGSIET
NFSSPLSIFSNPITAEATNFSDFDIRDIRKKPMSIYLGLTPDALITHEKIVNLFFSLLVNENCRELPEHN
PDLKYQCLILLDEFTSMGKSEVIERAVGFTAGYNLRFMFILQNEGQGQKSDMYGQEGWTTFTENSAVVLY
YPPKSKNALAKKISEEIGVRDMKISKRSISSGGGKGGSSRTRNDDVIERPVLLPEEIVSLRDKKNKARNI
AIREIITSEFSRPFIANKIIWFEEPEFKRRVDIARNNHVDIPNLFTQEVMDEIAKIAEIYLPKAGGKKVM
VAGGNVITNPDLDNHDKTDVSE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 24295..47114
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTH65_RS00110 (pMCR_1410_00024) | 19635..20167 | - | 533 | Protein_21 | thermonuclease family protein | - |
| HTH65_RS00115 (pMCR_1410_00025) | 20197..20850 | - | 654 | WP_000572551 | hypothetical protein | - |
| HTH65_RS00120 (pMCR_1410_00026) | 20862..21986 | - | 1125 | WP_000486719 | site-specific integrase | - |
| HTH65_RS00125 (pMCR_1410_00027) | 22399..23643 | - | 1245 | WP_000750517 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| HTH65_RS00130 (pMCR_1410_00028) | 23656..24291 | - | 636 | WP_000934979 | A24 family peptidase | - |
| HTH65_RS00135 (pMCR_1410_00029) | 24295..24723 | - | 429 | WP_001326801 | lytic transglycosylase domain-containing protein | virB1 |
| HTH65_RS00140 (pMCR_1410_00030) | 24843..25400 | - | 558 | WP_000095048 | type 4 pilus major pilin | - |
| HTH65_RS00145 (pMCR_1410_00031) | 25445..26554 | - | 1110 | WP_000974903 | type II secretion system F family protein | - |
| HTH65_RS00150 (pMCR_1410_00032) | 26545..28083 | - | 1539 | WP_000466225 | GspE/PulE family protein | virB11 |
| HTH65_RS00155 (pMCR_1410_00033) | 28108..28602 | - | 495 | WP_000912553 | type IV pilus biogenesis protein PilP | - |
| HTH65_RS00160 (pMCR_1410_00034) | 28586..29908 | - | 1323 | WP_000454142 | type 4b pilus protein PilO2 | - |
| HTH65_RS00165 (pMCR_1410_00035) | 29947..31590 | - | 1644 | WP_001035589 | PilN family type IVB pilus formation outer membrane protein | - |
| HTH65_RS00170 (pMCR_1410_00036) | 31583..32119 | - | 537 | WP_001220543 | sigma 54-interacting transcriptional regulator | virb4 |
| HTH65_RS00175 (pMCR_1410_00037) | 32166..34124 | - | 1959 | WP_000338976 | type IV secretory system conjugative DNA transfer family protein | - |
| HTH65_RS00180 (pMCR_1410_00038) | 34140..35195 | - | 1056 | WP_001542006 | P-type DNA transfer ATPase VirB11 | virB11 |
| HTH65_RS00185 (pMCR_1410_00039) | 35214..36353 | - | 1140 | WP_001542008 | TrbI/VirB10 family protein | virB10 |
| HTH65_RS00190 (pMCR_1410_00040) | 36343..37044 | - | 702 | WP_049824867 | TrbG/VirB9 family P-type conjugative transfer protein | - |
| HTH65_RS00195 (pMCR_1410_00041) | 37110..37844 | - | 735 | WP_000432282 | virB8 family protein | virB8 |
| HTH65_RS00200 (pMCR_1410_00043) | 38010..40367 | - | 2358 | WP_000548950 | VirB4 family type IV secretion system protein | virb4 |
| HTH65_RS00205 (pMCR_1410_00044) | 40373..40693 | - | 321 | WP_000362080 | VirB3 family type IV secretion system protein | virB3 |
| HTH65_RS00350 (pMCR_1410_00045) | 40764..41054 | - | 291 | WP_000865479 | conjugal transfer protein | - |
| HTH65_RS00215 (pMCR_1410_00046) | 41054..41638 | - | 585 | WP_065358197 | lytic transglycosylase domain-containing protein | virB1 |
| HTH65_RS00220 (pMCR_1410_00047) | 41659..42057 | - | 399 | WP_001153669 | hypothetical protein | - |
| HTH65_RS00225 (pMCR_1410_00048) | 42176..42613 | - | 438 | WP_034169416 | type IV pilus biogenesis protein PilM | - |
| HTH65_RS00230 (pMCR_1410_00049) | 42619..43854 | - | 1236 | WP_053899060 | toxin co-regulated pilus biosynthesis Q family protein | - |
| HTH65_RS00235 (pMCR_1410_00050) | 43857..44156 | - | 300 | WP_000835763 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| HTH65_RS00240 (pMCR_1410_00051) | 44204..45013 | + | 810 | WP_024237698 | DUF5710 domain-containing protein | - |
| HTH65_RS00245 (pMCR_1410_00052) | 45236..45460 | - | 225 | WP_000713562 | EexN family lipoprotein | - |
| HTH65_RS00250 (pMCR_1410_00053) | 45469..46113 | - | 645 | WP_001310442 | type IV secretion system protein | - |
| HTH65_RS00255 (pMCR_1410_00054) | 46119..47114 | - | 996 | WP_001028540 | type IV secretion system protein | virB6 |
| HTH65_RS00260 (pMCR_1410_00055) | 47118..47375 | - | 258 | WP_000739144 | hypothetical protein | - |
| HTH65_RS00265 (pMCR_1410_00056) | 47372..47725 | - | 354 | WP_223286767 | hypothetical protein | - |
| HTH65_RS00270 (pMCR_1410_00058) | 47945..48391 | - | 447 | WP_001243165 | hypothetical protein | - |
| HTH65_RS00275 (pMCR_1410_00059) | 48402..48572 | - | 171 | WP_000550720 | hypothetical protein | - |
| HTH65_RS00280 (pMCR_1410_00060) | 48576..49019 | - | 444 | WP_000964330 | NfeD family protein | - |
| HTH65_RS00285 (pMCR_1410_00061) | 49393..50346 | - | 954 | WP_072089442 | SPFH domain-containing protein | - |
| HTH65_RS00290 (pMCR_1410_00062) | 50373..50549 | - | 177 | WP_000753050 | hypothetical protein | - |
| HTH65_RS00295 (pMCR_1410_00063) | 50542..50757 | - | 216 | WP_001127357 | DUF1187 family protein | - |
| HTH65_RS00300 (pMCR_1410_00064) | 50750..51256 | - | 507 | WP_001326595 | CaiF/GrlA family transcriptional regulator | - |
Host bacterium
| ID | 14356 | GenBank | NZ_KU922754 |
| Plasmid name | pMCR_1410 | Incompatibility group | IncI2 |
| Plasmid size | 57059 bp | Coordinate of oriT [Strand] | 6374..6426 [-] |
| Host baterium | Kluyvera ascorbata strain WCH1410 |
Cargo genes
| Drug resistance gene | mcr-1.1 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |