Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 113901 |
| Name | oriT_pTM18-3 |
| Organism | Aeromonas sobria strain TM18 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_MF770242 (4322..4354 [-], 33 nt) |
| oriT length | 33 nt |
| IRs (inverted repeats) | 4..9, 13..18 (GCAAAC..GTTTGC) |
| Location of nic site | 27..28 |
| Conserved sequence flanking the nic site |
GTGCGCCCTT |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 33 nt
>oriT_pTM18-3
CACGCAAACGAAGTTTGCCTAAGTGCGCCCTTA
CACGCAAACGAAGTTTGCCTAAGTGCGCCCTTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file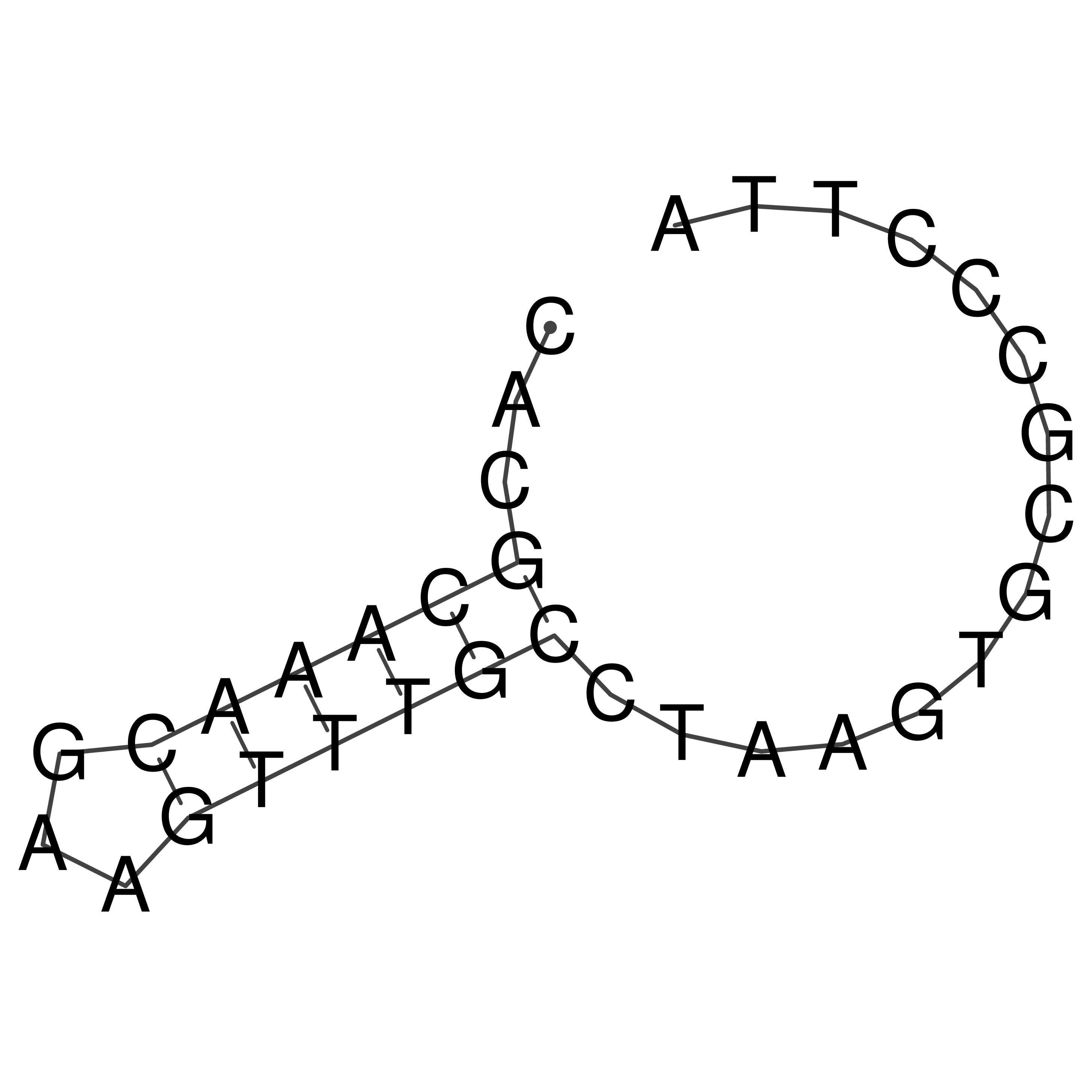
Relaxase
| ID | 8926 | GenBank | WP_172691897 |
| Name | mobQ_CJP16_RS00005_pTM18-3 |
UniProt ID | _ |
| Length | 514 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 514 a.a. Molecular weight: 57616.55 Da Isoelectric Point: 7.1156
>WP_172691897.1 MobQ family relaxase [Aeromonas sobria]
MAIYHCSVKTFSRRNGQSATAAAAYRAGEKLLDEQTGELHDYRKKGGVISATMILPSHAPLWASDRARLW
NEVERAETRINSTVAREFEIALPHELSPDDRERLAHEFARELVEEHGFAADVAIHAPPAFKKGDTGKMID
NQNHHAHILLSTRRLGPDGFTQKTREFDAKGRSDKASERFGPALIEYWRERFGVLQNQFLEAAGVEQKVD
HRSHQARGLAAEPTRHLGPAVVGFERRTGIDSEKRRSEARAVRAVDERVQALDRAARELAELDAQEVVIA
AELAENQRYVAMDDAELSDVISNAQPTPIDHQVASSWEVSQAKRAVTDLESKHDSVARQIESWEEGHSFK
TWLNSKGIGGAELDGLRTGLAQLGNRITKALGKVREVTEATHDRLWPEYNRRLTLFDELRELRQAKRQRK
ALQVEVAQSLEFSTIALQRSMGVLGWADGGKEWSACPKRLQTRIEDYLSTSGGEREALIAEIGRSVEVRD
LLARQKRGLDPAPEPERNRGPKLG
MAIYHCSVKTFSRRNGQSATAAAAYRAGEKLLDEQTGELHDYRKKGGVISATMILPSHAPLWASDRARLW
NEVERAETRINSTVAREFEIALPHELSPDDRERLAHEFARELVEEHGFAADVAIHAPPAFKKGDTGKMID
NQNHHAHILLSTRRLGPDGFTQKTREFDAKGRSDKASERFGPALIEYWRERFGVLQNQFLEAAGVEQKVD
HRSHQARGLAAEPTRHLGPAVVGFERRTGIDSEKRRSEARAVRAVDERVQALDRAARELAELDAQEVVIA
AELAENQRYVAMDDAELSDVISNAQPTPIDHQVASSWEVSQAKRAVTDLESKHDSVARQIESWEEGHSFK
TWLNSKGIGGAELDGLRTGLAQLGNRITKALGKVREVTEATHDRLWPEYNRRLTLFDELRELRQAKRQRK
ALQVEVAQSLEFSTIALQRSMGVLGWADGGKEWSACPKRLQTRIEDYLSTSGGEREALIAEIGRSVEVRD
LLARQKRGLDPAPEPERNRGPKLG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 14336 | GenBank | NZ_MF770242 |
| Plasmid name | pTM18-3 | Incompatibility group | - |
| Plasmid size | 4393 bp | Coordinate of oriT [Strand] | 4322..4354 [-] |
| Host baterium | Aeromonas sobria strain TM18 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |