Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 113795 |
| Name | oriT_p12TM-94 |
| Organism | Serratia marcescens strain 12TM |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CM008896 (18606..18710 [+], 105 nt) |
| oriT length | 105 nt |
| IRs (inverted repeats) | 32..37, 40..45 (GCCGGC..GCCGGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 105 nt
>oriT_p12TM-94
AGTACGGGACAAGATGTGTTTTTGGTACACCGCCGGCACGCCGGCAGTGACGCAAAAACGTCTTGGTCAGGGCAAGCCCCGACACCCCCTAACGAGGTTAGCTAT
AGTACGGGACAAGATGTGTTTTTGGTACACCGCCGGCACGCCGGCAGTGACGCAAAAACGTCTTGGTCAGGGCAAGCCCCGACACCCCCTAACGAGGTTAGCTAT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file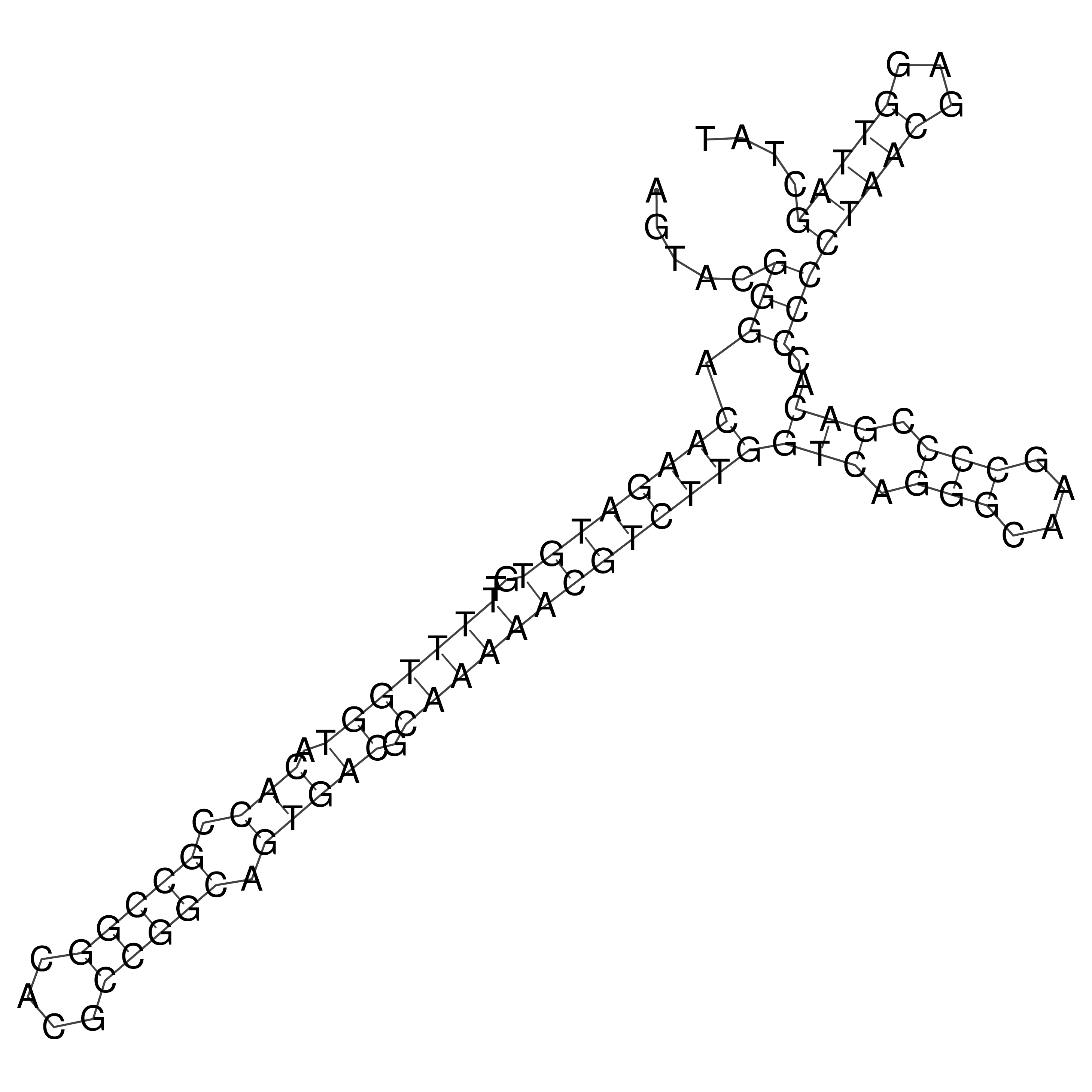
Relaxase
| ID | 8833 | GenBank | WP_101428087 |
| Name | traI_CS367_RS25795_p12TM-94 |
UniProt ID | _ |
| Length | 659 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 659 a.a. Molecular weight: 75087.88 Da Isoelectric Point: 10.0563
>WP_101428087.1 TraI/MobA(P) family conjugative relaxase [Serratia marcescens]
MVIPKIIEGRRDKKSSFGQLIKYMADKPSQELTDTVQPTPETALAVKSDMFEGLNNYLTRKQKVISTPVD
VEPGVQRVVVGDVTCQYNTFSLDGAAQEMNSVSQQSTRCKDPVMHYVLSWPDYEKPNDDQVFDSVKFTLA
SMGMSDHQYVAAIHRDTDNLHVHVAVNRINPQTYKAASSSFTKDTLHQACRLLELKNGWSHSNGAYVVND
RQQIVRNPHSKKERGNWRSLDRINKMENKEGVETLYRYIVGDEQVGGSRQNLIHVSAGLREAKSWDDVHK
TFAGIGLRVEKAQGKKGYVITHEHQNQKTAVKASLVFNKAQYTLKSMEERFGEYQPSHIEPAKVSVFKTA
YTPGAYRRDANKRLQRKIERAEERMLLKGRYRAYRNNLPIYSPDKDRIADEYRKIAQHTRLVKNNVRHSV
SDPHTRKLMYNLAEFKRLQAVANLRLSLREERNGFRAANPRLSYREWVEQEALKGDKAALSQMRGFAYSS
RKKEKYKQQLVEQIGFNRTFNAITSHDRDDVAVMASARHGVKPRLLKDGTVIFERDGKPVAADRGHIVLT
ESNGIDKEKTADLAIALTIAGKAKSVRVDGDGEFKELCCNRIVDAAVNHNHPVAQGITFTDAAQQAYAQN
EKHRLIREQNNSKNEMKLRSESDDKFNPK
MVIPKIIEGRRDKKSSFGQLIKYMADKPSQELTDTVQPTPETALAVKSDMFEGLNNYLTRKQKVISTPVD
VEPGVQRVVVGDVTCQYNTFSLDGAAQEMNSVSQQSTRCKDPVMHYVLSWPDYEKPNDDQVFDSVKFTLA
SMGMSDHQYVAAIHRDTDNLHVHVAVNRINPQTYKAASSSFTKDTLHQACRLLELKNGWSHSNGAYVVND
RQQIVRNPHSKKERGNWRSLDRINKMENKEGVETLYRYIVGDEQVGGSRQNLIHVSAGLREAKSWDDVHK
TFAGIGLRVEKAQGKKGYVITHEHQNQKTAVKASLVFNKAQYTLKSMEERFGEYQPSHIEPAKVSVFKTA
YTPGAYRRDANKRLQRKIERAEERMLLKGRYRAYRNNLPIYSPDKDRIADEYRKIAQHTRLVKNNVRHSV
SDPHTRKLMYNLAEFKRLQAVANLRLSLREERNGFRAANPRLSYREWVEQEALKGDKAALSQMRGFAYSS
RKKEKYKQQLVEQIGFNRTFNAITSHDRDDVAVMASARHGVKPRLLKDGTVIFERDGKPVAADRGHIVLT
ESNGIDKEKTADLAIALTIAGKAKSVRVDGDGEFKELCCNRIVDAAVNHNHPVAQGITFTDAAQQAYAQN
EKHRLIREQNNSKNEMKLRSESDDKFNPK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 10248 | GenBank | WP_254616853 |
| Name | t4cp2_CS367_RS26820_p12TM-94 |
UniProt ID | _ |
| Length | 326 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 326 a.a. Molecular weight: 37330.10 Da Isoelectric Point: 4.3073
>WP_254616853.1 MULTISPECIES: hypothetical protein [Enterobacterales]
MDDIFHNDRILIVMIPSLELSRGEAATLGRLYVTLQRMTISKDLGYQLEGKKEEVLLTHALNNQAPYGLI
YDELGQYFTSGMDTLSAQMRSLEKMGVFSSQDHPSLARGANGEVDSLIANTRVKYFESIEDRKTFEILRE
TVGQDYYSELSGQEAEHGTFSKTVREGDLYQIREKDRVNIRELRKQTEGQGVISFQDALVRSAAFYIPDN
EKFSSELPMRINRFIEVLPPSPATLYSIYPERKDDELAETNPDAMRFPPIKLFGYDKAANSLLEKIEVFY
ELQCGAISPEEMSVCLFELFAEWLDGTETDDVLYHQEDDLFPMIEM
MDDIFHNDRILIVMIPSLELSRGEAATLGRLYVTLQRMTISKDLGYQLEGKKEEVLLTHALNNQAPYGLI
YDELGQYFTSGMDTLSAQMRSLEKMGVFSSQDHPSLARGANGEVDSLIANTRVKYFESIEDRKTFEILRE
TVGQDYYSELSGQEAEHGTFSKTVREGDLYQIREKDRVNIRELRKQTEGQGVISFQDALVRSAAFYIPDN
EKFSSELPMRINRFIEVLPPSPATLYSIYPERKDDELAETNPDAMRFPPIKLFGYDKAANSLLEKIEVFY
ELQCGAISPEEMSVCLFELFAEWLDGTETDDVLYHQEDDLFPMIEM
Protein domains
No domain identified.
Protein structure
No available structure.
| ID | 10249 | GenBank | WP_257571886 |
| Name | t4cp1_CS367_RS26825_p12TM-94 |
UniProt ID | _ |
| Length | 382 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 382 a.a. Molecular weight: 43589.48 Da Isoelectric Point: 9.1360
>WP_257571886.1 hypothetical protein [Serratia marcescens]
MQQESVDSKKVLRPDGSFMEWFLTPNVQFGLLAILTVLGLFLPFTMLLSILILPPLMVAFTDRKFRPPLR
MPKDCEMFDDTLTTETQAEYRLGPIRIPHKVRERKKAKGILYVGYERGRLFGRELWLNMTDLLRHMVFFG
TTGSGKTETFYGFIVNFLLWCRGYCLSDGKADNKLAFATWSLARRFGREDDYYVLNLLTGSIDRFVNLVK
QESIPAQSNSVNLFSVAPPTFIIQLMESMLPQVGGDSAQWQDTAKAMMSALINALCYKRARGELLLSQRT
IQKNMSLPAMAALYVEAKKNGWHSEGYAALESYLENTPGFLLANAEYPETWEARAFEQHNYMSRQFLKTL
SLFNETYGHVFRKTAAISGWTIFSTMTAFSSS
MQQESVDSKKVLRPDGSFMEWFLTPNVQFGLLAILTVLGLFLPFTMLLSILILPPLMVAFTDRKFRPPLR
MPKDCEMFDDTLTTETQAEYRLGPIRIPHKVRERKKAKGILYVGYERGRLFGRELWLNMTDLLRHMVFFG
TTGSGKTETFYGFIVNFLLWCRGYCLSDGKADNKLAFATWSLARRFGREDDYYVLNLLTGSIDRFVNLVK
QESIPAQSNSVNLFSVAPPTFIIQLMESMLPQVGGDSAQWQDTAKAMMSALINALCYKRARGELLLSQRT
IQKNMSLPAMAALYVEAKKNGWHSEGYAALESYLENTPGFLLANAEYPETWEARAFEQHNYMSRQFLKTL
SLFNETYGHVFRKTAAISGWTIFSTMTAFSSS
Protein domains
No domain identified.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 21508..41936
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CS367_RS25775 (CS367_25705) | 16592..16903 | + | 312 | WP_004187333 | hypothetical protein | - |
| CS367_RS25780 (CS367_25710) | 17036..17572 | + | 537 | WP_004206920 | hypothetical protein | - |
| CS367_RS25785 (CS367_25715) | 18073..18438 | - | 366 | WP_015586046 | hypothetical protein | - |
| CS367_RS25790 (CS367_25720) | 18714..19031 | + | 318 | WP_004206923 | plasmid mobilization protein MobA | - |
| CS367_RS25795 (CS367_25725) | 19018..20997 | + | 1980 | WP_101428087 | TraI/MobA(P) family conjugative relaxase | - |
| CS367_RS25800 (CS367_25730) | 21011..21511 | + | 501 | WP_004206925 | DotD/TraH family lipoprotein | - |
| CS367_RS25805 (CS367_25735) | 21508..22287 | + | 780 | WP_004187315 | type IV secretory system conjugative DNA transfer family protein | traI |
| CS367_RS25810 (CS367_25740) | 22298..23461 | + | 1164 | WP_004206926 | plasmid transfer ATPase TraJ | virB11 |
| CS367_RS25815 (CS367_25745) | 23451..23711 | + | 261 | WP_004187310 | IcmT/TraK family protein | traK |
| CS367_RS26815 | 23736..27041 | + | 3306 | WP_227549828 | LPD7 domain-containing protein | - |
| CS367_RS26310 (CS367_25755) | 27007..27519 | + | 513 | WP_011091071 | hypothetical protein | traL |
| CS367_RS25825 (CS367_25760) | 27520..27732 | - | 213 | WP_305953721 | Hha/YmoA family nucleoid-associated regulatory protein | - |
| CS367_RS25830 (CS367_25765) | 27704..28114 | - | 411 | WP_004187465 | H-NS family nucleoid-associated regulatory protein | - |
| CS367_RS25835 (CS367_25770) | 28182..28895 | + | 714 | WP_223294958 | DotI/IcmL family type IV secretion protein | traM |
| CS367_RS25840 (CS367_25775) | 28904..30055 | + | 1152 | WP_015062843 | DotH/IcmK family type IV secretion protein | traN |
| CS367_RS25845 (CS367_25780) | 30067..31416 | + | 1350 | WP_004206932 | conjugal transfer protein TraO | traO |
| CS367_RS25850 (CS367_25785) | 31428..32132 | + | 705 | WP_004206933 | conjugal transfer protein TraP | traP |
| CS367_RS25855 (CS367_25790) | 32156..32686 | + | 531 | WP_004187478 | conjugal transfer protein TraQ | traQ |
| CS367_RS25860 (CS367_25795) | 32703..33092 | + | 390 | WP_011154472 | DUF6750 family protein | traR |
| CS367_RS25865 (CS367_25800) | 33138..33632 | + | 495 | WP_004187480 | hypothetical protein | - |
| CS367_RS25870 (CS367_25805) | 33629..36679 | + | 3051 | WP_101428088 | conjugal transfer protein | traU |
| CS367_RS25875 (CS367_25810) | 36676..37884 | + | 1209 | WP_004187486 | conjugal transfer protein TraW | traW |
| CS367_RS25880 (CS367_25815) | 37881..38531 | + | 651 | WP_060450166 | hypothetical protein | - |
| CS367_RS25885 (CS367_25820) | 38524..40524 | + | 2001 | WP_242447813 | DotA/TraY family protein | traY |
| CS367_RS25890 (CS367_25825) | 40569..41716 | + | 1148 | WP_088581786 | IS3 family transposase | - |
| CS367_RS26505 (CS367_25830) | 41781..41936 | + | 156 | WP_172448221 | hypothetical protein | traY |
| CS367_RS25900 (CS367_25835) | 41939..42592 | + | 654 | WP_004206935 | hypothetical protein | - |
| CS367_RS25905 (CS367_25840) | 42666..42896 | + | 231 | WP_004187496 | IncL/M type plasmid replication protein RepC | - |
| CS367_RS26890 | 43146..43196 | + | 51 | WP_031942489 | IncL/M-type plasmid replication leader peptide RepB | - |
| CS367_RS25910 (CS367_25845) | 43193..44248 | + | 1056 | WP_021526650 | plasmid replication initiator RepA | - |
| CS367_RS25915 (CS367_25850) | 45139..46347 | - | 1209 | Protein_60 | Tn3 family transposase | - |
Host bacterium
| ID | 14230 | GenBank | NZ_CM008896 |
| Plasmid name | p12TM-94 | Incompatibility group | IncL/M |
| Plasmid size | 80890 bp | Coordinate of oriT [Strand] | 18606..18710 [+] |
| Host baterium | Serratia marcescens strain 12TM |
Cargo genes
| Drug resistance gene | blaOXA-1, aac(6')-Ib-cr, aac(3)-IIa, blaCTX-M-15, sul2, dfrA14, aph(6)-Id |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |