Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 113730 |
| Name | oriT_FY|p2 |
| Organism | Serratia marcescens strain FY |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP053380 (59074..59123 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 19..25, 29..35 (GCAAAAT..ATTTTGC) |
| Location of nic site | 44..45 |
| Conserved sequence flanking the nic site |
GGTGTGATGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_FY|p2
ATTTAAATAGAAAAATCTGCAAAATACAATTTTGCGTGGGGTGTGATGCT
ATTTAAATAGAAAAATCTGCAAAATACAATTTTGCGTGGGGTGTGATGCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file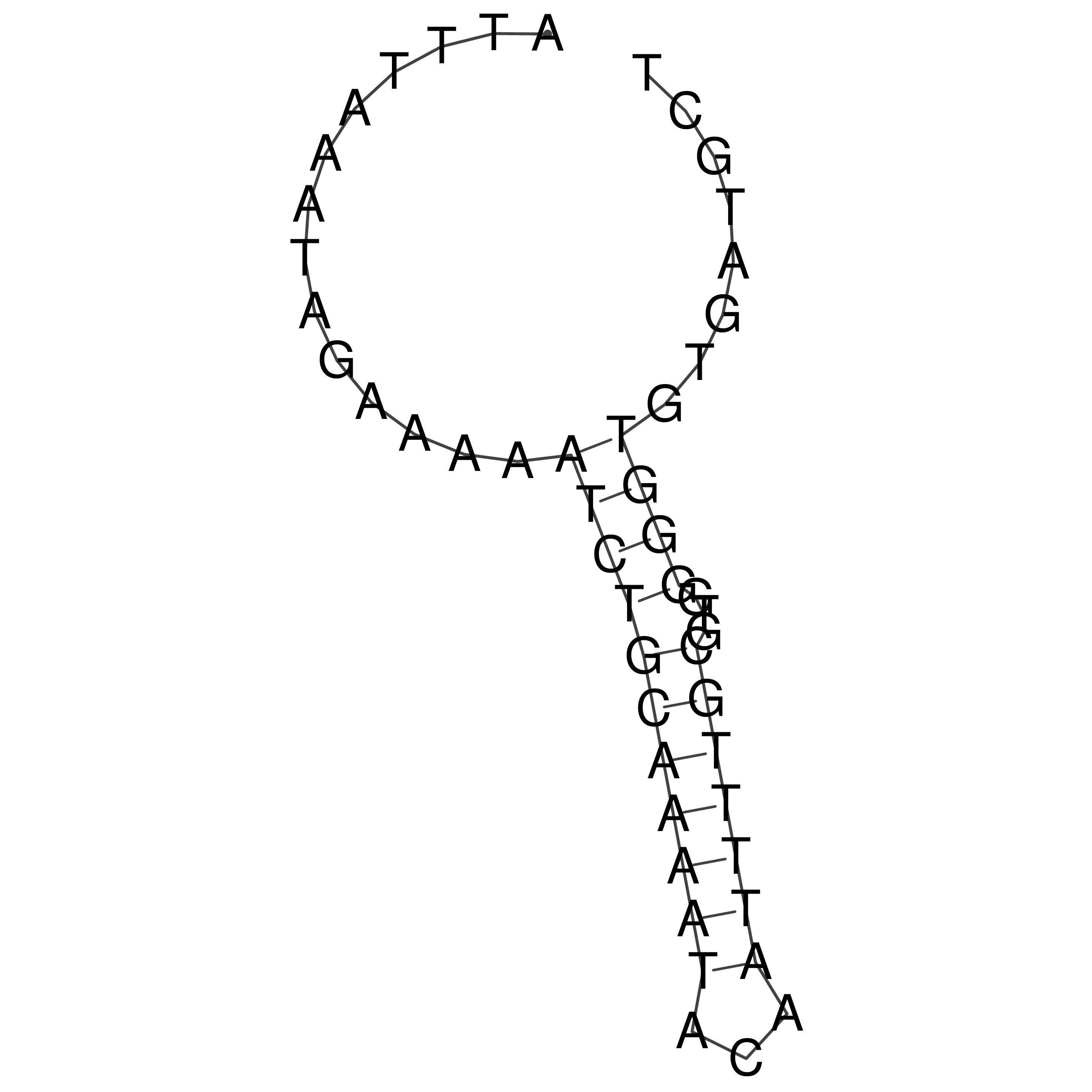
Relaxase
| ID | 8791 | GenBank | WP_171455178 |
| Name | traI_HMJ22_RS24605_FY|p2 |
UniProt ID | _ |
| Length | 1759 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1759 a.a. Molecular weight: 189025.78 Da Isoelectric Point: 5.8669
>WP_171455178.1 conjugative transfer relaxase/helicase TraI [Serratia marcescens]
MMSVWPVGPAGKAGDYYTHQDNYYVLGSMDERWVGQGAEALGLTGKVDVKDFVAVLEGKLPDGSDLTRMV
GGENKHRPGYDLTFSAPKSVSIMAMLGGDKRLIAAHNHAVEVAVREIEKMASTRVMEGGVSETKLTGNLV
VALFNHDTSRDLDPQLHTHAVVANVTQHDGKWQTLSSDTVGKTGFIENVYANQIAGGQIYRHILQREVEA
MGYETETTGPHGMWEFKRTEAVREPFSKRSHAIDAAAGPDASLKSRDMATLDTRKAKVASDPAALAVEWL
QEMKSAGFDAKAYLAEAQSRTMVTPSPTPLEAGGDALVDQAVGQAISALSDRQVNFTHNQVANKAISYLS
GEPGILDAVRDGIDRAIEREQLIPLDKEKGVFTSDIHLLDELSVAQLSRTHFDESAVVIGAPKADPRTGS
MPAYSDAVQQLAQHAPAMAVLSGLGGASVNRSRVAEVVELAREQGRDATVLTVDRQSQQYLQQSVHLDEG
RVVTLAALGSEALFAPNSTLVVDQAEKLTLKETVALLDGAVRNNVQLVLMDTEKRKGTGNALAVMKEAGV
PRYQFSGRQAVPTEVVSEPDKAQRFGQLAKGVVQALVEDRSAVAQVNGSADQRMLTAMIRDELKDTGLLG
REETHVATLTPVWLDSKNRGAIDTYREGMVMERWDAESRQRERFVIDRVGSQTRNLVLRNAEGEKHTLAV
KAVDKDWSLFRAGTMGVADGERLQVLGQEGRGKDALKAGTALTVVRAGAGELQVRVAGEPDATQKPADRA
LAVGSDVFSAVKVAPAYVESLGASVSSTAAVYAAVKKNELNSATLSGIARSGGHITLYSALDAHKTQEKV
AQHPAYRLAGEQVKQLAGEEGLDAAISGQKSGLFTPAQQAIHLAIPTLEGDSVAFGQAKLLAGSLDFAAK
GVSMEAIQTEIEAQVKRGELLKVDVAPGVGADLLVSRASFEAEKSIIRLIADGKDSMPALMSGTARNQLA
SIQSSLTQGQKESVDLILGTTDRFVAIQGYSGVGKTTQFKAVLAGIEQLPESERPRVVGLAPTHRAVGEM
QAAGVDAQTLASFLHDEGLKVASGEKPDYRNTLFVVDESSMVGNSDMARAYALIAEGNGRAVMSGDEAQL
EAIAPGAPFRLAQGRSAIDMAVMKEIVRQTPELKPAVYKMIENDVPGALDVVRTVAPANVPRQANAWVPA
KSVMELAVDIPDIHDAIVSDYLGRTEAARQDTLIITHLNADRHIINAGIHGERKAAGELKGESVSLPVLE
TANIRDGELRKLDTWARHRDAVVLLDNTYYRMADIDSQSRTVSLQDSEGNTRQFSPASAVREGVTLYNPR
TIEVNVGDRMRFSKSDVERGHVANSVWTVEAIKGDMVTLSDGQQTRTVNPREQQQDRHVDLAYAITAHGA
QGASERFAITLEGVEGGREPLVNRASAYVALSRAKEHVQVYTDNEAGWLNAIGKAKARTTAHDAVLGMAP
GSQKAAVDLYGKASSLSENALGRAVLRGSQLTGDTLAHYVSPGKKYPQPHAMLPMYDANGKPAGAWLAPL
ADTQGKSVGGLPTQGRVFGSDSAQFVALQGSRNNETRLAGTMQDGIRLAARYPDSGVVVRLQGDDVPFNP
QAMTGGRVWVNDETLKATARESGASDNVIKLPESPEERLQRELQTQADKLGREPNAPDPQRLAGLNDEQL
RALLARGELRAEDVKDRLPEAVMREVMGQDVAQRVDAAVRQVAREPVSTRSRLQQDEHNLVRDTHAEREL
NKEKTLGGD
MMSVWPVGPAGKAGDYYTHQDNYYVLGSMDERWVGQGAEALGLTGKVDVKDFVAVLEGKLPDGSDLTRMV
GGENKHRPGYDLTFSAPKSVSIMAMLGGDKRLIAAHNHAVEVAVREIEKMASTRVMEGGVSETKLTGNLV
VALFNHDTSRDLDPQLHTHAVVANVTQHDGKWQTLSSDTVGKTGFIENVYANQIAGGQIYRHILQREVEA
MGYETETTGPHGMWEFKRTEAVREPFSKRSHAIDAAAGPDASLKSRDMATLDTRKAKVASDPAALAVEWL
QEMKSAGFDAKAYLAEAQSRTMVTPSPTPLEAGGDALVDQAVGQAISALSDRQVNFTHNQVANKAISYLS
GEPGILDAVRDGIDRAIEREQLIPLDKEKGVFTSDIHLLDELSVAQLSRTHFDESAVVIGAPKADPRTGS
MPAYSDAVQQLAQHAPAMAVLSGLGGASVNRSRVAEVVELAREQGRDATVLTVDRQSQQYLQQSVHLDEG
RVVTLAALGSEALFAPNSTLVVDQAEKLTLKETVALLDGAVRNNVQLVLMDTEKRKGTGNALAVMKEAGV
PRYQFSGRQAVPTEVVSEPDKAQRFGQLAKGVVQALVEDRSAVAQVNGSADQRMLTAMIRDELKDTGLLG
REETHVATLTPVWLDSKNRGAIDTYREGMVMERWDAESRQRERFVIDRVGSQTRNLVLRNAEGEKHTLAV
KAVDKDWSLFRAGTMGVADGERLQVLGQEGRGKDALKAGTALTVVRAGAGELQVRVAGEPDATQKPADRA
LAVGSDVFSAVKVAPAYVESLGASVSSTAAVYAAVKKNELNSATLSGIARSGGHITLYSALDAHKTQEKV
AQHPAYRLAGEQVKQLAGEEGLDAAISGQKSGLFTPAQQAIHLAIPTLEGDSVAFGQAKLLAGSLDFAAK
GVSMEAIQTEIEAQVKRGELLKVDVAPGVGADLLVSRASFEAEKSIIRLIADGKDSMPALMSGTARNQLA
SIQSSLTQGQKESVDLILGTTDRFVAIQGYSGVGKTTQFKAVLAGIEQLPESERPRVVGLAPTHRAVGEM
QAAGVDAQTLASFLHDEGLKVASGEKPDYRNTLFVVDESSMVGNSDMARAYALIAEGNGRAVMSGDEAQL
EAIAPGAPFRLAQGRSAIDMAVMKEIVRQTPELKPAVYKMIENDVPGALDVVRTVAPANVPRQANAWVPA
KSVMELAVDIPDIHDAIVSDYLGRTEAARQDTLIITHLNADRHIINAGIHGERKAAGELKGESVSLPVLE
TANIRDGELRKLDTWARHRDAVVLLDNTYYRMADIDSQSRTVSLQDSEGNTRQFSPASAVREGVTLYNPR
TIEVNVGDRMRFSKSDVERGHVANSVWTVEAIKGDMVTLSDGQQTRTVNPREQQQDRHVDLAYAITAHGA
QGASERFAITLEGVEGGREPLVNRASAYVALSRAKEHVQVYTDNEAGWLNAIGKAKARTTAHDAVLGMAP
GSQKAAVDLYGKASSLSENALGRAVLRGSQLTGDTLAHYVSPGKKYPQPHAMLPMYDANGKPAGAWLAPL
ADTQGKSVGGLPTQGRVFGSDSAQFVALQGSRNNETRLAGTMQDGIRLAARYPDSGVVVRLQGDDVPFNP
QAMTGGRVWVNDETLKATARESGASDNVIKLPESPEERLQRELQTQADKLGREPNAPDPQRLAGLNDEQL
RALLARGELRAEDVKDRLPEAVMREVMGQDVAQRVDAAVRQVAREPVSTRSRLQQDEHNLVRDTHAEREL
NKEKTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 10190 | GenBank | WP_171455181 |
| Name | traD_HMJ22_RS24620_FY|p2 |
UniProt ID | _ |
| Length | 732 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 732 a.a. Molecular weight: 81631.31 Da Isoelectric Point: 5.1369
>WP_171455181.1 type IV conjugative transfer system coupling protein TraD [Serratia marcescens]
MSFNAKDMTQGGQIAFMRLRMFTQIGNLIFYCLFIAFFLVAICTLLWRVSIQNLTNGLIYWWVWMVEGWP
SISPSETLYHLDYYGKTLTYTAKQVLLDRYVSYCGQLIWREGIFACIVAAVACVTGFVVTVWILGRQGKQ
QSEDEITGGRTLTDNPAVVANMLKRDGMASDIIIDGLPLLKNSEVKNFCAMGSVGTGKSRLYRKLMDYAR
KRGDMVIVYDKSCDFVKDYYDPQHDKILNPQDARCVAWDLWSECLTLPDFENVADILIPMGSNEDPFWQG
SARTIFVEAAYRMRNDEDRSYAKLLNTLLSIDLPRLRAYLEGTPAANLVDGKIEKTAISIRSVLTNYAKA
LRYMQGVEKLGAPFTIRDWMRGVKDNGKNGWLFLTSDSASHKALKPLISMWLSIASHSLLSMGENPDRRV
WIFADEVPTLHKLPDLVSIVPEARKFGGCFVLGFQTYSQLEDIYGVKAAATLFGVLRTRFLFSVPDRVGA
EFAAGEVGEKEYRKASEQFSYGADPVRDGVSVGKDLERMQLVSYSDIQTLPDLCCYVTLPGPYPAVRLQM
VYKKVRQVAHPYEPRKLDAAADGRLVSAIEMREAETGNMDALFAATPILPAASARETADAPVSAVSKVPT
ENPASDVPVVSAATTATGGQEAELKQKPAPGDDDAEREELPPGISGDGEVVDIAQYEAWQAQQQVMGQQA
MQRREEANITIDRHRRELAEVHHGDSEPGDEF
MSFNAKDMTQGGQIAFMRLRMFTQIGNLIFYCLFIAFFLVAICTLLWRVSIQNLTNGLIYWWVWMVEGWP
SISPSETLYHLDYYGKTLTYTAKQVLLDRYVSYCGQLIWREGIFACIVAAVACVTGFVVTVWILGRQGKQ
QSEDEITGGRTLTDNPAVVANMLKRDGMASDIIIDGLPLLKNSEVKNFCAMGSVGTGKSRLYRKLMDYAR
KRGDMVIVYDKSCDFVKDYYDPQHDKILNPQDARCVAWDLWSECLTLPDFENVADILIPMGSNEDPFWQG
SARTIFVEAAYRMRNDEDRSYAKLLNTLLSIDLPRLRAYLEGTPAANLVDGKIEKTAISIRSVLTNYAKA
LRYMQGVEKLGAPFTIRDWMRGVKDNGKNGWLFLTSDSASHKALKPLISMWLSIASHSLLSMGENPDRRV
WIFADEVPTLHKLPDLVSIVPEARKFGGCFVLGFQTYSQLEDIYGVKAAATLFGVLRTRFLFSVPDRVGA
EFAAGEVGEKEYRKASEQFSYGADPVRDGVSVGKDLERMQLVSYSDIQTLPDLCCYVTLPGPYPAVRLQM
VYKKVRQVAHPYEPRKLDAAADGRLVSAIEMREAETGNMDALFAATPILPAASARETADAPVSAVSKVPT
ENPASDVPVVSAATTATGGQEAELKQKPAPGDDDAEREELPPGISGDGEVVDIAQYEAWQAQQQVMGQQA
MQRREEANITIDRHRRELAEVHHGDSEPGDEF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 10191 | GenBank | WP_079450624 |
| Name | traC_HMJ22_RS24700_FY|p2 |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 98864.80 Da Isoelectric Point: 5.7461
>WP_079450624.1 type IV secretion system protein TraC [Serratia marcescens]
MNLLDKVTHTANSLLSALKMPDEAAAANKVLGDMNFPQLSSVLPYRDYDEESGLFINAGSIGFVLEALPL
AGANEQVVLTLESLLRTKLPRGIALSVHLMASKRVGDLVESGLRDFSWSGKDADKFNAITRAFYLMAAEG
RFSLPPGVDMPLTLRNYRVFISYCVKSKRRNKALITELENLVKVLRASLMGAKIPTESLNDEDFINVVGE
IINHDPSQLYNKRRKLDPLKDLNFQCIDNGFGMDVYPDCVKINLRDAGSKKGSVARVMNFQLEQNPEMAF
LWNTADNYGNLLYPELSIATPFIITLTLVVEEQTKTSFEANRKYIDLEKKSRTSYAKFFPGTLEEMKEWG
DLRQSLASNQTAIVSFFYNVTVFCEDNDSVALRCEQDVINSYRKNGFDLISPRFKQHWNFLACLPFMAEH
GVFQDLAMHGAVHRAKTLNVVNLMPIVADNRLAPSGLLAPTYRNQLAFIDIYSEVMRNTNYNMAVCGTSG
AGKTGLIQPMLRSVLDTGGAAWVFDMGDGYKSLCENMGGVYIDAQTLKFNPFANVKDINLSATQIANLIS
VMASPDGNLDEVHESLLEMAVKAAWLSKQKNARIDDVRDFLRDAMDEEVYRDSPTIRNRLHEMTILLDKY
CSDGTYGEFFNSDEPSLRDDVNMVVLEMGGLKNRPDLLVAVMFSLMIYIEGRMYQTSRQSKKLCVIDEGW
KLLNFKNAKAGEFIEEGYRTARRHRGAYITITQNIKDFDSDTASSAAKAAWGNSSYKIILMQDASEFKTY
LQKNPDNFTQLQQDMIGKFESAKDQWFSSFMLMVSGKVSWHRLFVDPLSRAMYSSQGQDFEFMQARREQG
VSVHDAVYELAHRNFGEEMARIERWVEENYTGRAAA
MNLLDKVTHTANSLLSALKMPDEAAAANKVLGDMNFPQLSSVLPYRDYDEESGLFINAGSIGFVLEALPL
AGANEQVVLTLESLLRTKLPRGIALSVHLMASKRVGDLVESGLRDFSWSGKDADKFNAITRAFYLMAAEG
RFSLPPGVDMPLTLRNYRVFISYCVKSKRRNKALITELENLVKVLRASLMGAKIPTESLNDEDFINVVGE
IINHDPSQLYNKRRKLDPLKDLNFQCIDNGFGMDVYPDCVKINLRDAGSKKGSVARVMNFQLEQNPEMAF
LWNTADNYGNLLYPELSIATPFIITLTLVVEEQTKTSFEANRKYIDLEKKSRTSYAKFFPGTLEEMKEWG
DLRQSLASNQTAIVSFFYNVTVFCEDNDSVALRCEQDVINSYRKNGFDLISPRFKQHWNFLACLPFMAEH
GVFQDLAMHGAVHRAKTLNVVNLMPIVADNRLAPSGLLAPTYRNQLAFIDIYSEVMRNTNYNMAVCGTSG
AGKTGLIQPMLRSVLDTGGAAWVFDMGDGYKSLCENMGGVYIDAQTLKFNPFANVKDINLSATQIANLIS
VMASPDGNLDEVHESLLEMAVKAAWLSKQKNARIDDVRDFLRDAMDEEVYRDSPTIRNRLHEMTILLDKY
CSDGTYGEFFNSDEPSLRDDVNMVVLEMGGLKNRPDLLVAVMFSLMIYIEGRMYQTSRQSKKLCVIDEGW
KLLNFKNAKAGEFIEEGYRTARRHRGAYITITQNIKDFDSDTASSAAKAAWGNSSYKIILMQDASEFKTY
LQKNPDNFTQLQQDMIGKFESAKDQWFSSFMLMVSGKVSWHRLFVDPLSRAMYSSQGQDFEFMQARREQG
VSVHDAVYELAHRNFGEEMARIERWVEENYTGRAAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 36280..59677
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HMJ22_RS24610 | 32692..32919 | + | 228 | WP_171455179 | toxin-antitoxin system antitoxin VapB | - |
| HMJ22_RS24615 | 32919..33317 | + | 399 | WP_171455180 | tRNA(fMet)-specific endonuclease VapC | - |
| HMJ22_RS24620 | 33353..35551 | - | 2199 | WP_171455181 | type IV conjugative transfer system coupling protein TraD | - |
| HMJ22_RS24625 | 35541..35750 | - | 210 | WP_033641525 | hypothetical protein | - |
| HMJ22_RS24630 | 35753..36283 | - | 531 | WP_171455182 | hypothetical protein | - |
| HMJ22_RS24635 | 36280..39111 | - | 2832 | WP_171455183 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HMJ22_RS24640 | 39113..40480 | - | 1368 | WP_171455184 | conjugal transfer pilus assembly protein TraH | traH |
| HMJ22_RS24645 | 40477..40893 | - | 417 | WP_171455185 | hypothetical protein | - |
| HMJ22_RS24650 | 40886..41425 | - | 540 | WP_171455186 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HMJ22_RS24655 | 41412..41726 | - | 315 | WP_171455187 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HMJ22_RS24660 | 41737..42462 | - | 726 | WP_253718574 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HMJ22_RS24665 | 42528..44432 | - | 1905 | WP_171455189 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HMJ22_RS24670 | 44578..45252 | - | 675 | WP_253718565 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HMJ22_RS24675 | 45263..46261 | - | 999 | WP_171455190 | conjugal transfer pilus assembly protein TraU | traU |
| HMJ22_RS24680 | 46258..46635 | - | 378 | WP_171455191 | hypothetical protein | - |
| HMJ22_RS24685 | 46647..47285 | - | 639 | WP_171455229 | type-F conjugative transfer system protein TraW | traW |
| HMJ22_RS24690 | 47282..48163 | - | 882 | WP_171455192 | DsbA family protein | - |
| HMJ22_RS24695 | 48247..48666 | - | 420 | WP_171455193 | type-F conjugative transfer system protein TrbI | - |
| HMJ22_RS24700 | 48663..51293 | - | 2631 | WP_079450624 | type IV secretion system protein TraC | virb4 |
| HMJ22_RS24705 | 51307..51846 | - | 540 | WP_171455230 | type IV conjugative transfer system lipoprotein TraV | traV |
| HMJ22_RS24710 | 51871..52257 | - | 387 | WP_171455194 | hypothetical protein | - |
| HMJ22_RS24715 | 52254..52472 | - | 219 | WP_070914249 | hypothetical protein | virb4 |
| HMJ22_RS24720 | 52469..53068 | - | 600 | WP_171455195 | hypothetical protein | - |
| HMJ22_RS24725 | 53065..54459 | - | 1395 | WP_171455196 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HMJ22_RS24730 | 54459..55187 | - | 729 | WP_171455197 | type-F conjugative transfer system secretin TraK | traK |
| HMJ22_RS24735 | 55177..55740 | - | 564 | WP_147882578 | type IV conjugative transfer system protein TraE | traE |
| HMJ22_RS24740 | 55751..56056 | - | 306 | WP_042785939 | type IV conjugative transfer system protein TraL | traL |
| HMJ22_RS24745 | 56068..56433 | - | 366 | WP_070914241 | type IV conjugative transfer system pilin TraA | - |
| HMJ22_RS24750 | 56547..56744 | - | 198 | WP_094141473 | TraY domain-containing protein | - |
| HMJ22_RS24755 | 56921..57505 | + | 585 | WP_094141474 | helix-turn-helix domain-containing protein | - |
| HMJ22_RS24760 | 57496..58332 | + | 837 | WP_171455198 | queuosine precursor transporter | - |
| HMJ22_RS24765 | 58402..58788 | - | 387 | WP_171455199 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HMJ22_RS24770 | 59207..59677 | + | 471 | WP_171455200 | transglycosylase SLT domain-containing protein | virB1 |
| HMJ22_RS24775 | 59962..60258 | - | 297 | WP_171455201 | DUF5983 family protein | - |
| HMJ22_RS24780 | 60360..61187 | - | 828 | WP_171455202 | DUF932 domain-containing protein | - |
| HMJ22_RS24785 | 61244..61615 | - | 372 | WP_171455203 | dehydrogenase | - |
| HMJ22_RS24790 | 62152..62382 | - | 231 | WP_171455157 | hypothetical protein | - |
| HMJ22_RS24795 | 62339..62566 | - | 228 | WP_171455204 | hypothetical protein | - |
| HMJ22_RS24800 | 62633..62920 | - | 288 | WP_004929861 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| HMJ22_RS24805 | 62917..63168 | - | 252 | WP_038883487 | type II toxin-antitoxin system Phd/YefM family antitoxin | - |
| HMJ22_RS24810 | 63620..64345 | - | 726 | WP_171455205 | plasmid SOS inhibition protein A | - |
Host bacterium
| ID | 14165 | GenBank | NZ_CP053380 |
| Plasmid name | FY|p2 | Incompatibility group | - |
| Plasmid size | 87789 bp | Coordinate of oriT [Strand] | 59074..59123 [+] |
| Host baterium | Serratia marcescens strain FY |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |