Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 113602 |
| Name | oriT_S17BD05944|p39 |
| Organism | Shigella sonnei strain S17BD05944 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP099774 (34421..34512 [-], 92 nt) |
| oriT length | 92 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 92 nt
>oriT_S17BD05944|p39
CGGGGTGTCGGGGCTAAGCCCTGACCAGGTGGTAATCGTATAGCCGTGCGTGCGCGGTTATACGATTACACATCCTGTCCCGTTTTTCAGGC
CGGGGTGTCGGGGCTAAGCCCTGACCAGGTGGTAATCGTATAGCCGTGCGTGCGCGGTTATACGATTACACATCCTGTCCCGTTTTTCAGGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file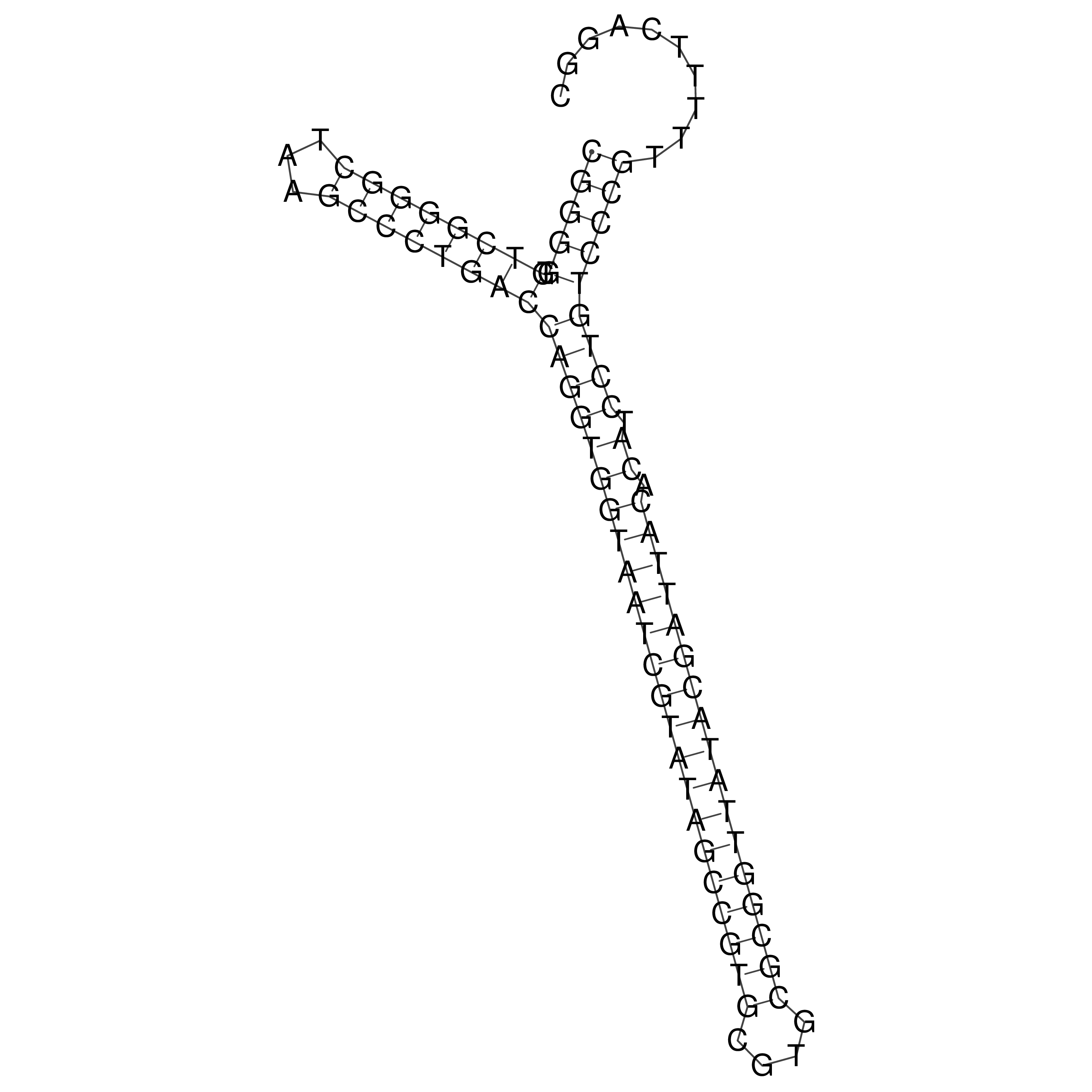
Relaxase
| ID | 8711 | GenBank | WP_044810581 |
| Name | Relaxase_NHV82_RS00230_S17BD05944|p39 |
UniProt ID | _ |
| Length | 906 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 906 a.a. Molecular weight: 104926.61 Da Isoelectric Point: 8.3624
>WP_044810581.1 MULTISPECIES: relaxase/mobilization nuclease domain-containing protein [Enterobacteriaceae]
MNAIIPKKRRDGKSSFEDLIAYTSVRDDVPEDELRPDTEMKAEIPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCLSLESAAEEMQKAAELAHFSKDDTDPVFHYILSWPAHESPRSEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINCLPWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIDQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGV
LVVKDGWDKAREGIALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVGTPGRYDPEAINVDIRPEKVAETE
SLKQYACRHFAERLPAMARNGELKSSLAVHRTLAEAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVVPGERFRGHSIGSQPVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
GLIQCRSLIEEMIADGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAAADIFERVKPECRYNPELAASDEVEPGFRRDPEIRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAYIRVQFRDPQLRKLHYHIAEVQRMQALIRLKESVKEERLSLIAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNVDRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHTDRNELAEKLNLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTITRPDVDLHRQRSEQYYREYIRQQTRLSESHDDNYTLRQEKTWEPPSPGM
MNAIIPKKRRDGKSSFEDLIAYTSVRDDVPEDELRPDTEMKAEIPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCLSLESAAEEMQKAAELAHFSKDDTDPVFHYILSWPAHESPRSEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINCLPWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIDQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGV
LVVKDGWDKAREGIALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVGTPGRYDPEAINVDIRPEKVAETE
SLKQYACRHFAERLPAMARNGELKSSLAVHRTLAEAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVVPGERFRGHSIGSQPVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
GLIQCRSLIEEMIADGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAAADIFERVKPECRYNPELAASDEVEPGFRRDPEIRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAYIRVQFRDPQLRKLHYHIAEVQRMQALIRLKESVKEERLSLIAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNVDRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHTDRNELAEKLNLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTITRPDVDLHRQRSEQYYREYIRQQTRLSESHDDNYTLRQEKTWEPPSPGM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 5095 | GenBank | WP_001291057 |
| Name | WP_001291057_S17BD05944|p39 |
UniProt ID | A0A142CND4 |
| Length | 110 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 110 a.a. Molecular weight: 12596.69 Da Isoelectric Point: 10.7335
>WP_001291057.1 MULTISPECIES: plasmid mobilization protein MobA [Enterobacteriaceae]
MSEKKTRSGSEKRQKNVLIAVRFSPEEAEIVKEKAEKNGLTVSTLIRKTVLGKQINARIDEDFLKELMRL
GRLQKHLFVEGKRTGDKEYAEVLVAITELANTLRRNLMGR
MSEKKTRSGSEKRQKNVLIAVRFSPEEAEIVKEKAEKNGLTVSTLIRKTVLGKQINARIDEDFLKELMRL
GRLQKHLFVEGKRTGDKEYAEVLVAITELANTLRRNLMGR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A142CND4 |
T4CP
| ID | 10086 | GenBank | WP_000077662 |
| Name | trbC_NHV82_RS00235_S17BD05944|p39 |
UniProt ID | _ |
| Length | 766 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 766 a.a. Molecular weight: 86964.85 Da Isoelectric Point: 6.5117
>WP_000077662.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Enterobacteriaceae]
MSQHHVNPDLIHRTAWGNPLWNALHNLNITGLCLAGSIITALIWPLALPVCLLFTLVTSVIFMLQRWRCP
LRMPMTLSLDDPSQDRKVRRSLFSFWPTLFQYEADETFPARGIFYVGYRRINDIGRELWLSMDDLTRHIM
FFATTGGGKTETTFAWLLNPLCWGRGFTFVDGKAQNDTTRTIWYLSRRFGREDDIEVINFMNGGKSRSEI
IQSGEKSRPQSNTWNPFAFSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDMSLVRTPSAWTEEPRKQHAYLSGQFSE
TFTTFAETFGDVFAADAGDIDIRDSIHSDRILIVMIPALDTSAHTTSALGRMFVTQQSMLLARDLGYRLE
GTDAQTLEVKKYKGSFPYICILDEVGAYYTDRIAVEATQVRSLEFSLIMTAQDQERIEGQTSATNTATLM
QNAGTKFAGRIVSDDKTARTVKNAAGEEARARMGSLQRHDGVMGESWVDGNQITIQMESKIDVQDLIRLN
AGEFFTVFQGDVVPSASVYIPDSEKSCDSDPVVINRYISVEAPRLERLRRLVPRTVQRRLPTPEHVSSII
GVLTAKPSRKRRKNRTEPYRIIDTFQRQLATAQTSLDLLPQYDTDIESRANELWKKAVHTINNTTREERR
VCYITLNRPEEEHSGPEDIPSVKVILNTLLPLEILLPVAENNKSQTINNDHPQKSCGAQPTTDEKY
MSQHHVNPDLIHRTAWGNPLWNALHNLNITGLCLAGSIITALIWPLALPVCLLFTLVTSVIFMLQRWRCP
LRMPMTLSLDDPSQDRKVRRSLFSFWPTLFQYEADETFPARGIFYVGYRRINDIGRELWLSMDDLTRHIM
FFATTGGGKTETTFAWLLNPLCWGRGFTFVDGKAQNDTTRTIWYLSRRFGREDDIEVINFMNGGKSRSEI
IQSGEKSRPQSNTWNPFAFSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDMSLVRTPSAWTEEPRKQHAYLSGQFSE
TFTTFAETFGDVFAADAGDIDIRDSIHSDRILIVMIPALDTSAHTTSALGRMFVTQQSMLLARDLGYRLE
GTDAQTLEVKKYKGSFPYICILDEVGAYYTDRIAVEATQVRSLEFSLIMTAQDQERIEGQTSATNTATLM
QNAGTKFAGRIVSDDKTARTVKNAAGEEARARMGSLQRHDGVMGESWVDGNQITIQMESKIDVQDLIRLN
AGEFFTVFQGDVVPSASVYIPDSEKSCDSDPVVINRYISVEAPRLERLRRLVPRTVQRRLPTPEHVSSII
GVLTAKPSRKRRKNRTEPYRIIDTFQRQLATAQTSLDLLPQYDTDIESRANELWKKAVHTINNTTREERR
VCYITLNRPEEEHSGPEDIPSVKVILNTLLPLEILLPVAENNKSQTINNDHPQKSCGAQPTTDEKY
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 40093..81318
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| NHV82_RS00235 | 37812..40112 | - | 2301 | WP_000077662 | F-type conjugative transfer protein TrbC | - |
| NHV82_RS00240 | 40093..41217 | - | 1125 | WP_000152380 | DsbC family protein | trbB |
| NHV82_RS00245 | 41214..42539 | - | 1326 | WP_024186660 | IncI1-type conjugal transfer protein TrbA | trbA |
| NHV82_RS00510 | 43336..43431 | + | 96 | WP_001363141 | DinQ-like type I toxin DqlB | - |
| NHV82_RS00255 | 43494..43670 | - | 177 | WP_001054907 | hypothetical protein | - |
| NHV82_RS00260 | 43820..44287 | - | 468 | WP_000598962 | thermonuclease family protein | - |
| NHV82_RS00265 | 44447..45070 | + | 624 | WP_001155250 | helix-turn-helix domain-containing protein | - |
| NHV82_RS00270 | 45268..45483 | - | 216 | WP_000266544 | hypothetical protein | - |
| NHV82_RS00275 | 45487..45855 | - | 369 | WP_001302696 | hypothetical protein | - |
| NHV82_RS00280 | 46154..46396 | - | 243 | WP_001363144 | hypothetical protein | - |
| NHV82_RS00285 | 46726..46878 | + | 153 | WP_001335079 | Hok/Gef family protein | - |
| NHV82_RS00290 | 47016..48602 | - | 1587 | WP_000004561 | TnsA endonuclease N-terminal domain-containing protein | - |
| NHV82_RS00295 | 48876..49526 | - | 651 | WP_001044247 | plasmid IncI1-type surface exclusion protein ExcA | - |
| NHV82_RS00300 | 49615..51780 | - | 2166 | WP_052894952 | DotA/TraY family protein | traY |
| NHV82_RS00305 | 51855..52424 | - | 570 | WP_000650752 | conjugal transfer protein TraX | - |
| NHV82_RS00310 | 52421..53626 | - | 1206 | WP_001189548 | conjugal transfer protein TraW | traW |
| NHV82_RS00315 | 53584..54204 | - | 621 | WP_000286854 | hypothetical protein | traV |
| NHV82_RS00320 | 54204..57248 | - | 3045 | WP_001024766 | conjugal transfer protein | traU |
| NHV82_RS00325 | 57299..57550 | + | 252 | WP_052971984 | hypothetical protein | - |
| NHV82_RS00330 | 57543..58169 | - | 627 | WP_000785703 | hypothetical protein | traT |
| NHV82_RS00515 | 58276..58527 | - | 252 | WP_001277573 | hypothetical protein | - |
| NHV82_RS00340 | 58584..58982 | - | 399 | WP_000088813 | DUF6750 family protein | traR |
| NHV82_RS00345 | 59029..59559 | - | 531 | WP_000987021 | conjugal transfer protein TraQ | traQ |
| NHV82_RS00350 | 59556..60269 | - | 714 | WP_044810583 | conjugal transfer protein TraP | traP |
| NHV82_RS00355 | 60266..61603 | - | 1338 | WP_001272018 | conjugal transfer protein TraO | traO |
| NHV82_RS00360 | 61607..62581 | - | 975 | WP_000547385 | DotH/IcmK family type IV secretion protein | traN |
| NHV82_RS00365 | 62592..63287 | - | 696 | WP_001287545 | DotI/IcmL family type IV secretion protein | traM |
| NHV82_RS00370 | 63299..63649 | - | 351 | WP_001056367 | hypothetical protein | traL |
| NHV82_RS00375 | 63666..67727 | - | 4062 | WP_108433737 | LPD7 domain-containing protein | - |
| NHV82_RS00380 | 67791..68081 | - | 291 | WP_000817801 | IcmT/TraK family protein | traK |
| NHV82_RS00385 | 68078..69226 | - | 1149 | WP_000775210 | plasmid transfer ATPase TraJ | virB11 |
| NHV82_RS00390 | 69210..70046 | - | 837 | WP_000745049 | type IV secretory system conjugative DNA transfer family protein | traI |
| NHV82_RS00395 | 70043..70501 | - | 459 | WP_001170111 | DotD/TraH family lipoprotein | - |
| NHV82_RS00400 | 70605..71807 | - | 1203 | WP_021529228 | conjugal transfer protein TraF | - |
| NHV82_RS00405 | 71909..72730 | - | 822 | WP_044810585 | hypothetical protein | traE |
| NHV82_RS00410 | 72943..74067 | - | 1125 | WP_021511763 | site-specific integrase | - |
| NHV82_RS00415 | 74391..74546 | - | 156 | WP_001358489 | hypothetical protein | - |
| NHV82_RS00420 | 74904..75284 | - | 381 | WP_253075073 | pilus assembly protein | - |
| NHV82_RS00425 | 75565..76926 | - | 1362 | WP_052977226 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| NHV82_RS00430 | 76944..77570 | - | 627 | WP_044810571 | prepilin peptidase | - |
| NHV82_RS00435 | 77586..78071 | - | 486 | WP_001080767 | lytic transglycosylase domain-containing protein | virB1 |
| NHV82_RS00440 | 78116..78652 | - | 537 | WP_000111534 | type 4 pilus major pilin | - |
| NHV82_RS00445 | 78714..79808 | - | 1095 | WP_044810572 | type II secretion system F family protein | - |
| NHV82_RS00450 | 79810..81318 | - | 1509 | WP_044810573 | GspE/PulE family protein | virB11 |
| NHV82_RS00455 | 81421..81879 | - | 459 | WP_000095884 | type IV pilus biogenesis protein PilP | - |
| NHV82_RS00460 | 81869..83164 | - | 1296 | WP_044810574 | type 4b pilus protein PilO2 | - |
| NHV82_RS00465 | 83185..84804 | - | 1620 | WP_001035559 | PilN family type IVB pilus formation outer membrane protein | - |
| NHV82_RS00470 | 84836..85249 | - | 414 | WP_124983997 | type IV pilus biogenesis protein PilM | - |
Host bacterium
| ID | 14037 | GenBank | NZ_CP099774 |
| Plasmid name | S17BD05944|p39 | Incompatibility group | IncB/O/K/Z |
| Plasmid size | 92062 bp | Coordinate of oriT [Strand] | 34421..34512 [-] |
| Host baterium | Shigella sonnei strain S17BD05944 |
Cargo genes
| Drug resistance gene | blaCTX-M-15, blaTEM-1B |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |